InSeon Yoo
Proceedings of the 2004 ACM workshop on Visualization and data mining for computer security, pp.82-89
ISBN 1-58113-974-8
October 2004
 Download PDF (531.02Kb) (You need to be registered on forum)
Download PDF (531.02Kb) (You need to be registered on forum)Permission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage and that copies bear this notice and the full citation on the first page. To copy otherwise, to republish, to post on servers or to redistribute to lists, requires prior specific permission and/or a fee.
VizSEC/DMSEC'04, October 29, 2004, Washington, DC, USA. Copyright 2004 ACM 1-58113-974-8/04/0010 ...$5.00.
Telecommunications, Networks and Security Research Group,This paper concentrates on visualizing computer viruses without using virus specific signature information as a prior stage of the very important problem of detecting computer viruses. In this paper, we address the fact that each viruses have its own character to be distinguished although it is inserted in the executable file. They cannot hide their own feature through the SOM visualization; this feature is like a DNA to determine an individual's unique genetic code. We present how virus codes affect the whole program projection. Without each virus signature, we present how the virus pattern in Windows executable files tells us their family. We show that the variant of each virus also can be covered with each virus mask, which is produced by SOM. We also present the file structure based SOMs of Windows executable files.
K.6.5 [Management of Computing and Information Systems]: Security and Protection - Invasive software; I.2.6 [Artificial Intelligence]: Learning - Connectionism and neural nets; I.5.2 [Pattern Recognition]: Design Methodology - Pattern analysis
Security
Visualization, Windows Executable Viruses, Self-Organizing Maps
The classic virus-detection techniques look for the presence of a virus-specific sequence of instructions, called a virus signature, inside the program: if the signature is found, it is highly probable that the program is infected. For example, the Win95.CIH (Chernobyl) virus is detected by checking for the hexadecimal sequence like following [7]:
E800 0000 005B 8D4B 4251 5050 0F01 4C24 FE5B 83C3 1CFA 8B2B
We aim to design the SOM in a way that neurons will flag the presence of peculiar patterns in Windows executable files and that the position of the active neurons will reflect the position of potentially malicious content in the file. We expect that we can find a similar pattern in files infected by viruses of the same family. Although anti-virus software needs to detect variants of each virus with different virus signature, our approach shows us that each virus family has a virus mask1 like a DNA. We will present how virus codes affect the SOM projection of the whole Windows executable program and show the results of considering several Windows viruses in this paper.
Self-Organizing Map (SOM) [2] is an unsupervised neural network method which has properties of both vector quantization and vector projection algorithms. A SOM consists of neurons organized on a regular low-dimensional grid (Figure 1). Each neuron is a d-dimensional weight vector (prototype vector, codebook vector) where d is equal to the dimension of the input vectors. The neurons are connected to adjacent neurons by a neighbourhood relation, which dictates the topology, or structure, of the map.
The SOM training algorithm includes the best-matching weight vector, and its topological neighbours on the map, which are updated: the region around the best-matching vector is stretched towards the presented training sample. The end result is that the neurons on the grid become ordered: neighbouring neurons have similar weight vectors. In the traditional sequential training, samples are presented to the map one at a time, and the algorithm gradually moves the weight vectors towards them. In the batch training, the data set is presented to the SOM as a whole, and the new weight vectors are weighted averages of the data vectors.
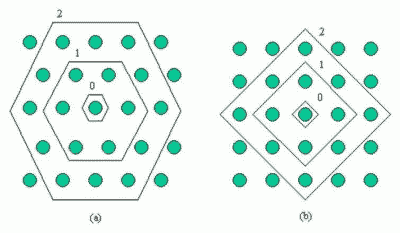
Figure 1: Discrete neighbourhood (size 0, 1, and 2) of the centermost unit:(a) hexagonal lattice, (b) rectangular lattice. The innermost polygon corresponds to 0-neighbourhood, the second to the 1-neighbourhood and the biggest to the 2-neighbourhood.
SOM combines vector quantization and vector projection. The goal of SOM is to create a topologically ordered mapping of the data in the sense of a discredited principal surface or curve.
The SOM has properties of both vector quantization and vector projection algorithms. The quantization from the N training samples to M prototypes reduces the original data set to a smaller, but still representative, set to work with. Further analysis is performed primarily, or at least initially using the prototype vectors instead of all of the data. Using the reduced data set is only valid if it really is representative of the original data. When the number of prototypes approaches infinity and neighbourhood width is very large, numerical experiments have shown that the results are relatively accurate even for a small number of prototypes [3]. While the connection between the density of prototypes of SOM and the input data has not been derived in the general case, it can be assumed that the SOM roughly follows the density of the training data.
To be able to visualize the prototypes efficiently, vector projection is needed. Together the set of prototype vectors and their projections form a low-dimensional map of the data manifold. Since the prototype vectors of the SOM have well-defined positions on the low-dimensional map grid, the SOM is a kind of vector projection algorithm. The projection of a data sample can be defined to be the index b or location rb of its BMU (the best matching unit) on the map grid. As a projection algorithm, SOM has an important advantage over many other methods. The topological ordering of map units depends primarily on the local neighbourhood, which is defined on the map grid. Since there are more map units where data density is high, the neighbourhood in these areas becomes smaller as measured in the input space. Thus, the projection tunes to local data density.
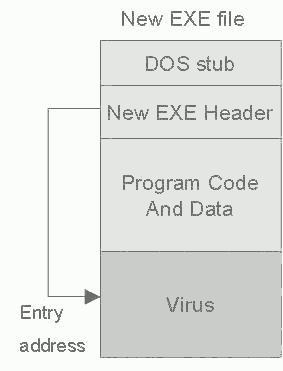
Figure 2: Virus positions in New EXE file.
The most common method of virus infection is by appending the virus to the end of file. In this process the virus changes the top of the file in such a way that the virus code is executed first. This kind of appending is simple and usually effective. The virus writer does not need to know anything about the program to which the virus will append and the appended program simply serves as a carrier for the virus [6].
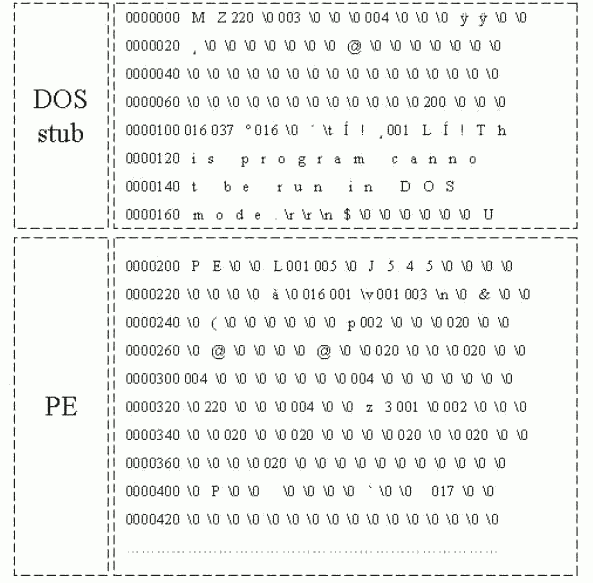
Figure 3: Windows Executable file: DOS Stub and PE header part
In the Windows executables (NewEXE - NE, PE, LE, LX), the fields in the NewEXE header are changed and the virus code is appending to the end of the program code. The structure of this header is much more complicated and there are more fields to be changed; the starting address, the number of sections in the file, properties of the sections etc. In addition to that, before infection, the size of the file may increase to a multiple of one paragraph (16 bytes) in DOS or to a section in Windows. The size of the section depends on the properties of the EXE file header. Figure 2 shows this case.
According to these 4 different areas in virus-infected files, we worked out to train SOMs. The real data of the virus-infected file is like in Figure 3 and Figure 4. In these figures, the location part is counted by octal numbering. When we examined several virus-infected files, most files have the same size of DOS stub (say, 128 bytes), and the other parts are flexible. In addition, apart from virus code, only PE header part is filled with quite similar pattern, which contains text, data, source and relocation.
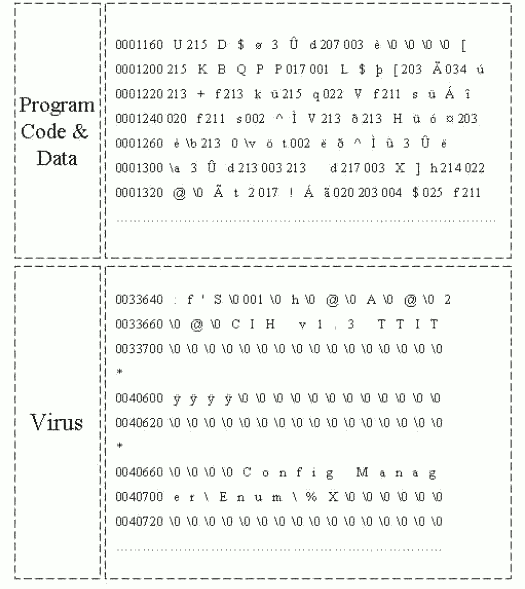
Figure 4: Windows Executable file: Program Code & Data and Virus position.
Furthermore, the program code & data part is filled with encrypted characters which are made by certain compilers. However, from virus code part, the characters look quite different from original program codes. As we examine like in Figure 3 and Figure 4, virus part character feature is different from the other program codes, which means that once program codes were compiled, the virus code were inserted by force. Since we have found these differences, we attempted to label each part, such as DS for the DOS stub, PE for the NewEXE header, PR for the program code & data, and VS for the virus code, to see which part is displayed differently by SOMs. After training and testing several virus-infected files, we have strong confidence that this virus part is inserted by force, and this virus code should have different code scheme compared with original program code, since the original code is compiled by a certain compiler. [Table 1] is our test data file information. As you see [Table 1], most of files have the same DOS stub size.
| Virus file | PE | PR | VS |
|---|---|---|---|
| Win95.CIH Ver 1.2 | 128 | 576 | 11632 |
| Win95.CIH Ver 1.3 | 128 | 624 | 14256 |
| Win95.CIH Ver 1.4 | 128 | 576 | 1968 |
| Win95.Boza.A | 128 | 1024 | 2016 |
| Win95.Boza.C | 256 | 1536 | 3072 |
| Win32.Apparition | 128 | 1024 | 38912 |
| Win32.HLLP.Semisoft | 128 | 1024 | 41360 |
Table 1: location starting point information of Test files (Unit: bytes)
Note. DOS stub always starts from 0000. PE : PE header, PR: Program Code & Data, VS: Virus Code
Viruses are called polymorphic if they cannot, or can but with great difficulty be detected using the virus signature. This is achieved by two main ways. The first is by encrypting the main code of the virus with non-constant key with random sets of decryption commands, the second is by changing the executable virus code. Polymorphic viruses exist of all kinds from boot and file, even macro viruses. In this paper, we are not dealing with these polymorphic viruses separately, because these polymorphic viruses can be included in parasitic viruses or macro viruses. Especially, changing executable codes is mostly by macro viruses, which randomly change the names of their variables, insert empty lines or change their codes in some other ways while making copies of themselves. Therefore the operating algorithm of a virus remains unchanged, but the virus code changes virtually completely from one infection to another.
We assume polymorphic and metamorphic viruses are somehow inserted in the executable files. Thus their figures, whether they are encrypted or not, are also distinguished compared with the other program codes.
To train and visualize SOMs from virus-infected files, we use SOM Toolbox 2.0 [1], a software library for MATLAB 5.3 [4]. The projection is that the originally high-dimensional reference vector space is compressed into two dimensions, making the visualization of the data possible. Unified distance matrix (u-matrix), is a method of displaying SOMs. It represents the map as a regular grid of neurons. The size and topology of the map can readily be observed from the picture where each element represents a neuron. First, when generating a u-matrix, a distance matrix between the reference vectors of adjacent neurons of two-dimensional map is formed. Then, some representation for the matrix is selected, e.g., a grey-level image. The colours in the figure have been selected so that in the black/white printout, the darker the colour between two neurons is, the closer is the relative distance between them. (In colour printout, blue colour part means closeness of distance between two neurons.) In addition, labelling is used to categorize the units or some units by giving them names. We use several Windows executable files to test. [Table 2] is the file information.
| variants of the virus | file size |
|---|---|
| Before infection by CIH1.2 | 11,632 |
| Before infection by CIH1.3 | 14,256 |
| Before infection by CIH1.4 | 1,968 |
| Win95.CIH Ver 1.2 | 19,536 |
| Win95.CIH Ver 1.3 | 36,864 |
| Win95.CIH Ver 1.4 | 4,608 |
| Before infection by Win95.Boza.A | 2,016 |
| Before infection by Win95.Boza.C | 3,072 |
| Win95.Boza.A | 12,408 |
| Win95.Boza.C | 16,384 |
| Before infection by Win32.Apparition | 38,912 |
| Win32.Apparition | 96,239 |
| Before infection by Win32.HLLP.Semisoft | 41,360 |
| Win32.HLLP.Semisoft | 59,904 |
Table 2: Test file information (Unit: Bytes)
To train SOMs, we make our data like a table. Each row of the table is one data sample. The columns of the table are the variables of the data set. The items on the row are the variables, or components, of the data set (Figure 5). The variables might be the properties of an object, or a set of measurements measured at a specific time. Every sample has the same set of variables. Thus each column of the table holds all values for one variable.
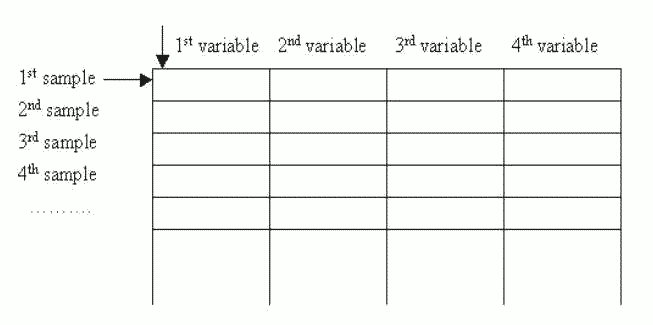
Figure 5: Table-format data: there can be any number of samples, but all samples have fixed length, and consist of the sample variables.
To match this table structure, we transformed binary format of virus-infected files with each 4 bytes for each column variables, e.g. each sample contains 32 bytes of virus-infected files. In addition, to deal with binary data efficiently, we converted all data into unsigned integer format. So through SOM normalization, this integer data can be normalized between 0 and 1.
The Win95.CIH (Chernobyl) [5] is a Windows95/98/NT specific parasitic virus infecting Windows PE (Portable Executable) files, about 1Kbyte of length. There are three original virus versions (1.2, 1.3 and 1.4) known, which are very closely related and only differ in few parts of their code. They have different lengths, texts inside the virus code and trigger date.
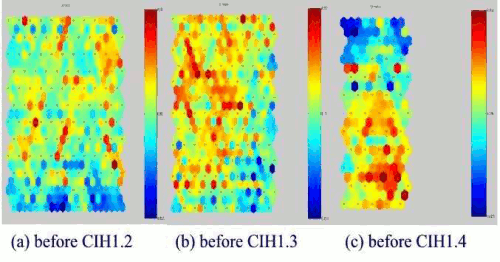
Figure 6: SOMs of Windows EXE files before Win95.CIH virus infection.
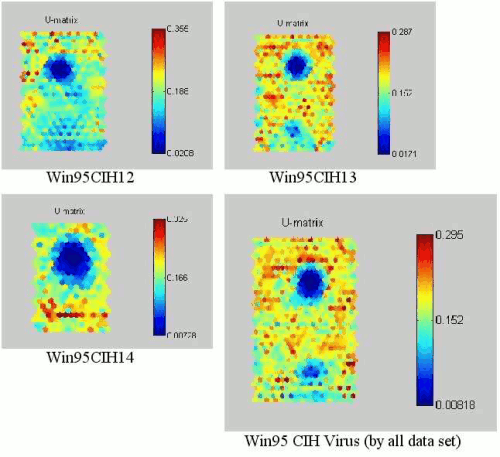
Figure 7: SOMs of NEW EXE files infected by Win95.CIH viruses.
Figure 6 shows the test Windows executable files before Win95.CIH virus infected. As you can see all the SOM of the test Windows executable files are different from each other. Figure 7 shows the trained SOMs of Win95.CIH 1.2, 1.3 and 1.4 infected test Windows executable files. Each Win95.CIH (Chernobyl) virus has obvious location (the upper of centre) of lower degree of weight data2. Although each Windows executable file is different, the SOM projection of CIH virus-infected files look similar and have same sort of projection map. We could call this similar sort of figure in each virus as a virus mask. Hence, this can be called CIH virus mask. Anti-virus software needs to detect this Win95.CIH virus with different virus signature in each version. However, our approach tells us that these are same family like having a same DNA.
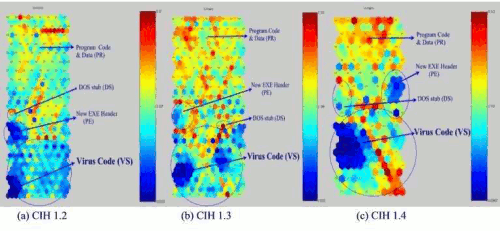
Figure 8: Virus Distribution of CIH 1.2, 1.3, and 1.4 virus.
To check that the upper centre part is filled with the CIH virus code, we trained the SOM with label which categorized by given name which we gave according to the [Table 1]. We made [Table 1] based on the file structure and when we made the data set, we put the label on each column. The result of the projection with labels is like in Figure 8. This projection is based on the labels, therefore, the whole figure (Figure 8) is different from previous figures (Figure 7).
We add round signs in each area e.g. DS (DOS stub), PE (New EXE Header), VS (Virus Code) area except PR (Program Code & Data) area. Since PR (Program Code & Data) area is been big enough, we do not need to make a distinguished round for this. As Figure 8 is shown, there are two part has smaller distances than the other parts, e.g., PE and VS. This tells us that PE and VS includes smaller likelihood in their code. Even if PE also has smaller distance between two neurons, VS has major of dark colour in black/white printout, (or blue colour in colour printout).
Win95 Boza virus is the first known virus infecting Windows Portable Executable (PE) files, such files are used by Windows 95 and Windows NT. However, Boza does not infect machines running the Microsoft Windows NT operating system. Boza's spreading technique resembles some of the early DOS viruses. When the first DOS viruses were found in 1980's, they were very simple compared to some of the currently known polymorphic multipartite fast infecting stealth viruses. However, it is not a dangerous parasitic NewEXE (PE) virus. It searches for EXE files, checks the files for PE signature, then creates new section named ".vlad", and writes its code into that section.
We trained two different Boza virus files. Figure 9 shows us the Boza virus mask. In addition, the lighter the colour between two neurons is, the smaller is the relative distance between them. We assume that the major of lighter colour in the upper centre has virus codes. To check and prove this, we made another projection with labels like in Figure 10. As we expected, the major of smaller likelihood part is the Boza virus code. Although NewEXE header code also has smaller likelihood, it does not change the majority.
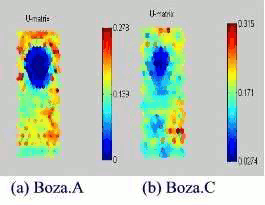
Figure 9: SOMs of Win95 Boza.A and Boza.C viruses.
This is a memory resident Windows32 (Windows95/NT) parasitic infector. The virus has a very unusual structure. The main part (about 60K) is the virus code (virus routines and C runtime library), text strings, icon and other data used by the virus while installing and spreading. The next block (3.5K) contains a packed (with LZ method) MS Word template - Word macro virus. The third block (21K) contains packed (by LZ) virus source code. And the last block (3K) contains resources file that is used when the virus runs Borland C compiler.
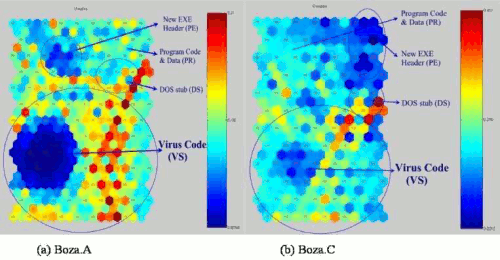
Figure 10: Virus Distribution of Boza.A and Boza.C viruses.
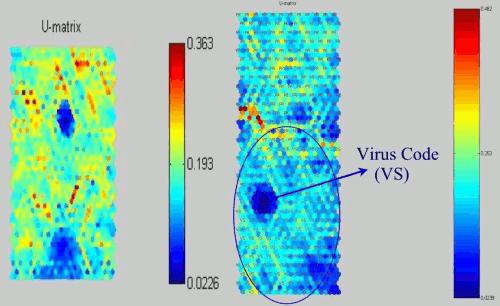
Figure 11: SOMs of Win NT apparition virus and its distribution.
As we looked the previous virus SOMs, this SOM also has the Win32.apparition virus mask. Figure 11 shows the projection of Win32.apparition virus and the other projection with label for the distribution. In addition, since this virus code part has unusual structure, the distribution itself causes VS part is quite similar with the PR parts. Nevertheless, the major of the smaller likelihood part is virus code.
Win32.HLLP.Semisoft virus is an unusual file infector which infects files under Windows 95 and Windows NT. The virus creates these 59,904 byte files in the WINDOWS directory: WINIPX.EXE, WINIPXA.EXE, WINSRVC.EXE, and EXPLORE.EXE. The virus infects other EXE files as they are executed once the virus is loaded into memory. The virus depends on a network card being installed to work fully. The virus may have been intended as a prototype of a "spy" program that would intercept information and send this out via a TCP/IP connection. A task "6.666" interferes with normal shutdown. The infected files all have a Notepad Icon when they are visible in Explorer.
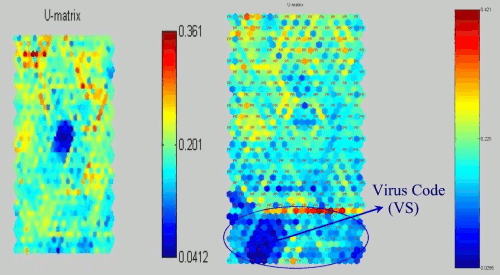
Figure 12: SOMs of Win32 HLLP.Semisoft virus and its distribution.
| CIH1.2 | CIH1.3 | CIH1.4 | Boza.A | Boza.C | apparition | HLLP.Semisoft | |
|---|---|---|---|---|---|---|---|
| Quantization error | 0.393 | 0.393 | 0.393 | 0.206 | 0.377 | 0.445 | 0.461 |
| Topographic error | 0.055 | 0.050 | 0.050 | 0.041 | 0.015 | 0.108 | 0.127 |
| Error Percentage | 14.6764 | 20.6107 | 20.6107 | 13.5314 | 29.3233 | 10.2605 | 28.9616 |
Figure 12 shows the projection of Win32.HLLP.Semisoft virus and the other projection with label for the distribution. As we looked the previous virus SOMs, this SOM also has the Win32.HLLP.Semisoft virus mask and the major of the smaller likelihood part is virus code.
After the SOM has been trained, it is important to know whether it has properly adapted itself to the training data. Because it is obvious that one optimal map for the given input data must exist, several map quality measures have been proposed. Usually, the quality of the SOM is evaluated based on the mapping precision and the topology preservation.
The mapping precision measure describes how accurately the neurons respond to the given data set. For example, if the reference vector of the BMU calculated for a given testing vector xi is exactly the same xi, the error in precision is then 0. Normally, the number of data vectors exceeds the number of neurons and the precision error is thus always different from 0.
A common measure that calculates the precision of the mapping is the average quantization error over the entire data set:
1 n
Eq = --- ∑ ||xi + mc||
N i=1
Where, x is a sample vector and m is a reference vector.
The topology preservation measure describes how well the SOM preserves the topology of the studied data set. Unlike the mapping precision measure, it considers the structure of the map. For a strangely twisted map, the topographic error is big even if the mapping precision error is small. A simple method for calculating the topographic error:
1 n
Et = --- ∑ u(xk)
N k=1
Where, u(xk) is 1 if the first and second BMUs of xk are not next to each other. Otherwise u(xk) is 0.
[Table 3] summarizes error ratios of our test virus files. There are three items in the Table, a quantization error is for mapping precision, a topographic error is for topology preservation, and error percentage is for the label error per distance (similarity). The lower quantization error is, the more exact the neuron responds. In addition, the lower topographic error is, the better the SOM preserves the topology.
Although there are variants of the virus-infected files, SOM projections tell us that they are a same family because of their own virus mask. It is like having a same DNA in the same family. We believe, using this virus mask, we can apply to find out variant viruses, which are changed some part from original virus codes.
We investigated the Windows virus-infected files, in fact, the NewEXE file format, using SOMs. As we expected, we have found a virus pattern in a same virus-infected files. Although anti-virus software needs to detect variants of each virus with different virus signatures, our approach shows us that each virus family has a virus mask like a DNA. Initial experiments show that this approach appears to be successful. In addition, we shall attempt to find out virus pattern similarity, not only a same virus family, but also a virus mask based on the virus-infected file structure in the future. How to detect computer virus using this virus mask? This is the next step of this ongoing project, and this could be a future work.
1 We have found sort of distinguished virus sign or special feature in the SOM reflection. We call this a virus mask.
2 The darker the colour between two neurons is, the closer is the relative distance between them. The upper is of the bar, the bigger is the relative distance between data. In black/white printout, the bar displays similar colour of black in the SOM. However, most of black colour in the SOMs represents the blue colour part or close distance between two neurons, which means the black colour part represents that the relative distance is smaller.
[Back to index] [Comments (0)]