Sanjay Goel, Stephen Bush
2003
To appear in the Complexity Journal, vol. 9, issue 2, Nov-Dec 2003, Special Issue: "Resilient & Adaptive Defense of Computing Networks. Sanjay Goel is a professor at the University at Albany, SUNY, e-mail: goel@albany.edu. Stephen F. Bush is with the GE Global Research Center, Niskayuna, NY, 12309, USA phone: 518-387-6827; fax: 518-387-4042; e-mail: bushsf@research.ge.com.
This paper presents results in two mutually complementary areas: distributed immunological information assurance and a new signature matching technique based upon Kolmogorov Complexity. This paper introduces a distributed model for security based on biological paradigms of Epidemiology and Immunology. In this model each node in the network has an immune system that identifies and destroys pathogens in the incoming network traffic as well as files resident on the node. The network nodes present a collective defense to the pathogens by working symbiotically and sharing pathogen information with each other. Each node compiles a list of pathogens that are perceived as threats by using information provided from all the nodes in the network. The signatures for these pathogens are incorporated into the detector population of the immune systems to increase the probability of detection. Critical to the success of this system is the detection scheme, which should not only be accurate but also efficient. Three separate schemes for detecting pathogens are examined, namely, Contiguous string matching, Hamming Distance, and Kolmogorov Complexity. This work provides a model of the system and examines the efficiency of different detection schemes. A simulation model is built to study the sensitivity of model parameters, such as signature length, sampling rate, network topology, etc. to detection of pathogens.
Immunology, Epidemiology, Information Assurance, Kolmogorov Complexity, Signature Matching.
Advances in computer security are emerging continuously yet, the number of computer crimes continue to rise unabated. The government, industry, and home users alike are taking serious steps to make computers and networks more secure, however, hackers seem to be unaffected by these measures and have unfettered access to any computer and data on the Internet. Viruses spread rampantly across networks in spite of the presence of Intrusion Detection Systems (IDS) because small changes to viruses render the IDS systems ineffective. Perpetrating computer crimes is no longer the domain of the perceived computer geniuses but of average users who use recipes and manuals from the Internet and follow simple steps to perpetrate these attacks. According to a CERT report the number of reported crimes has gone up from 6 in 1998 to over 82,000 in 2002 [11].
It is not clear whether the security has progressively worsened over time or if the increased crime rates are a function of increased usage, awareness and reporting. It is clear, however, that the stakes in computer security are rising steadily as commerce grows on the Internet. A Jupiter Media Metrix [35] report claims that online retail sales (B2C) will reach $104 billion in 2005 and $130 billion by 2006, up from $34 billion in 2001. According to a Gartner Group report [34], B2B commerce will grow from $919 billion dollars in 2002 to 8.3 trillion dollars in year 2005. Some of the recent attacks on the Internet [14] have caused billions of dollars in losses, associated with loss of integrity of the data, cost of system reconfiguration, as well as loss of essential computing services for extended periods of time. Once an intrusion is detected significant resources are spent just in determining the source of intrusion, identifying systems that have been impacted and in patching all the systems to prevent future attacks. After each virus attack the cleanup activities include checking integrity of systems and data, patching operating system (or software) and updating virus definitions in detection systems.
Clearly, current measures have been ineffective in controlling the spread of viruses and in controlling intrusion as evidenced by the increasing rate of computer crime. Most of the existing security measures have been preventive, with a focus on perimeter defenses of the computer system. A proactive security model that focuses on identifying and neutralizing pathogens that invade a system is suggested to complement perimeter defenses. This paper presents a security model that takes inspiration from the biological paradigm of Immunology and creates a reactive computer security system that detects and neutralizes pathogens invading the system. This is certainly not the first security model based on an immunological paradigm. Kephart [40] and Forrest [23] have done seminal work on the use of immunological paradigms for computer security. Since then a large body of knowledge has emerged in the design of Artificial Immune Systems (AIS). Several applications have been developed using Artificial Immune Systems, such as, fraud detection, computer security, and robotic control. Some of these will be discussed in Section 3. The applications, though conceptually elegant, have not been practical to deploy on a large scale.
This paper discusses the extant literature on immunological paradigms for computer security and addresses fundamental limitations of these systems that render them intractable to practical implementation. Critical to the success of Immune System-based security models is a scheme for representation and detection of pathogens, since it strongly influences the computational complexity of the system. This paper explores different schemes for representing pathogen signatures and their efficacy in detecting pathogens. The investigation focuses on the comparison of traditional string matching techniques (i.e., hamming distance, contiguous string match) with the use of a novel matching technique, first proposed in this paper, based upon Kolmogorov Complexity. This paper proposes the concept of collective defense where nodes on the network share information and resources with their neighbors to provide a collective defense against pathogens, thereby relieving the computational burden on individual nodes. Concepts from Epidemiology and the Theory of Random Graphs are used to explore the conceptual underpinnings of the mechanism of collective defense. Kolmogorov Complexity has been proposed as a pervasive metric that transcends both node and network level defenses in the proposed model.
The paper is organized as follows. Section II discusses the biological immune system and the analogy to computer immune systems. Section III discusses relevant literature on Immunology, Epidemiology, Random Graphs, and Kolmogorov Complexity. Section IV presents the architecture of the immune system incorporating collective defense. In section V, a mathematical model for performance of the immune system is presented. In section VI, multiple schemes for the representation of virus signatures and pathogen detection are presented and compared using analytic and simulation based results. The paper concludes with a discussion of the findings and plans for future research.
The role of the human immune system is to protect our body from pathogens, such as, viruses, bacteria, microbes, etc.... The immune system consists of different kinds of cells, which operate autonomously and interact with each other to create complex chains of events leading to the destruction of pathogens. At a high level, cells can be categorized into two groups: detectors and effectors. The role of detectors is to identify pathogens and the role of effectors is to neutralize pathogens.
There are two kinds of immune responses evoked by the immune system: innate response and specific response. The innate immune response is the natural resistance of the body to foreign antigens and is non-specific towards invaders in the body. During this response, a specialized class of cells called phagocytes (macrophages and neutrophils) is used. These specialized cells, which have surface receptors that match many common bacteria, have remained unchanged through the evolutionary period [36]. This system reacts nearly instantly on detection of pathogens in the body, however, it is incapable of recognizing viruses and bacteria that mutate and evolve.
The innate immune response is complemented by the adaptive immune response, in which antibodies are generated to specific pathogens that are not recognized by the phagocytes. The adaptive response system uses lymphocytes, which unlike phagocytes that contain receptors for multiple strains have receptors for a specific strain. Lymphocytes are produced in the bone marrow, which generates variants of genes that encode the receptor molecules and mature in the thymus. When an antigen is encountered, it is presented to the lymphocytes in the lymphatic system. The lymphocytes that match proliferate by cloning and subsequently differentiate into B-cells that are used for generating antibodies and T-cells that destroy infected cells and activate other cells in the immune system. Most cells that proliferate to fight pathogens die; a few are converted into memory cells that retain the signature of the pathogen that was matched. This leads to a rapid response the next time a similar pathogen is encountered; this is the principle used in vaccinations and inoculations. Over a long period of disuse these memory cells die, as evidenced by weakening immunity to vaccinations over time. Lymphocytes have a fixed lifetime and if during this period they do not match a pathogen, they automatically die.
The key to the functioning of the immune system is the detection mechanism that is employed in the immune system. Recognition is based on pattern matching between complimentary protein structures of the antigen and the detector. The primary purpose of the genetic mechanism in the thymus and bone marrow is to generate proteins with different physical structures. The immune system recognizes pathogens by matching protein structure of the pathogen with that of the receptor. If the receptor of the antigen and the detector fit together like a three-dimensional jigsaw puzzle, a match is found.
A fundamental problem with the detection mechanism of the immune system is the computational complexity. For example, if there are 50 different attributes with four different values, over six million different detectors are required to cover the entire search space. The virus structures that can arise due to different protein configurations are virtually infinite. In spite of high efficiency in creating detectors and pattern matching at the molecular level, the problem of maintaining a detector for each possible pathogen protein structure is computationally infeasible. The human immune mechanism solves this problem by using generalizations in matching where some features of the
structure are ignored during the match. This is called specificity of match; the more the features are ignored during the match, the lower the specificity. The lower the specificity, the fewer is the number of detectors that are required for matching a population of pathogens and the more non-specific is the response. An explanation of specificity is elegantly described in John Holland's description of the classifier systems [29]. If d is the detector signature and p is the pathogen signature, such that, 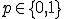 and
and 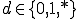 where * is the don't-care condition. The detector matches the pathogen when p and d contain a complimentary bit sequence, and specificity is defined as the number (or ratio) of the non-* values during the match. For instance p=11101010, d=11101010 shows a match with specificity of 1, while p=11101010, d = 11****10 shows a match with specificity of four. If the detector d = ******** then the specificity of the match would be zero and the detector would match any pathogen.
where * is the don't-care condition. The detector matches the pathogen when p and d contain a complimentary bit sequence, and specificity is defined as the number (or ratio) of the non-* values during the match. For instance p=11101010, d=11101010 shows a match with specificity of 1, while p=11101010, d = 11****10 shows a match with specificity of four. If the detector d = ******** then the specificity of the match would be zero and the detector would match any pathogen.
To cover the space of all possible non-self proteins the immune system uses detectors with low specificity. This enables the immune system to detect most pathogens with only a few detectors; however, it results in poor discrimination ability and a weak response to the pathogen intrusion. The immune system counters this problem by employing a process called affinity maturation that results in creation of detectors (B-Cells) with high specificity targeted towards the specific pathogens that are encountered. During the affinity maturation process, a B-cell that is activated clones itself using a high mutation rate (somatic hypermutation) to create a population of detectors that match the pathogen but maintain sufficient diversity to evolve more specific detectors. The cloned detectors are themselves activated and cloned with the cloning rate proportional to the fitness of the detector. The fitness function of the detectors is the affinity for the specific pathogen. The detectors thus compete with each other with the fitter detectors proliferating faster and creating more specific detectors. As a result of affinity maturation a very generic response is thus quickly transformed into a specific response.
Another problem that arises in the immune system is that since the cells of the human body are made of the same protein building blocks as the pathogens, the immune system not only has to identify foreign proteins but also needs to distinguish between legitimate cells that belong to the body from pathogens that invade the body. This process is called self/non-self discrimination. The immune system enforces this discrimination by exposing nascent lymphocytes to the protein structures of the legitimate cells in the human body and destroying those lymphocytes (clonal deletion) that match protein structures of legitimate cells in the human body, prior to activating them for potential exposure to pathogens. The rest of the lymphocytes are retained for a specific time period during which they have a chance of matching a pathogen and becoming permanent. Whenever this mechanism fails and the lymphatic system fails to destroy a lymphocyte that matches a self-cell, autoimmune diseases can ensue.
The T-Cells mature in the thymus through which most self-proteins circulate and are thus sensitized to the self-proteins of the body via clonal deletion. The B-Cells, though sensitized to self-proteins themselves, may generate clones during the affinity maturation process that are not tolerant to the self-proteins of the body. To ensure tolerance of B-cells to the self-proteins, costimulation is employed by the immune system. Thus, in order to be activated, B-Cells must receive a signal from two separate sources. The first signal occurs when the receptors of the B-cells match the protein structure of the foreign protein. The helper T-cells deliver the second signal when they recognize the proteins in the pathogen that was intercepted by the B-cell. In order to get a response from the helper T-cells, the B-cells ingest the pathogen and dissect it into protein fragments that are presented to the T-cells. The T-cells match the fragments with their receptors and if a match occurs send a signal to the B-cell to trigger a response.
Undoubtedly there is a strong analogy between a biological immune system and a computer immune system. A study of the human immune system reveals unique characteristics that can be abstracted for use in an artificial immune system, viz., self-organization, learning, memory, clonal selection, and pattern recognition. The computer immune systems proposed in the literature have tried to mimic the biological immune system very closely. This paper, on the other hand, focuses on specific impediments in the use of Immune Systems for creating computer security applications.
A major characteristic of biological immune systems is discrimination between self and non-self. This is critical to the human immune system, a closed system. This concept is less relevant to computer security systems that need to dynamically interact with other computers and devices on the network. The definition of self is not static, since network composition is changing continuously, with new nodes entering the network and existing nodes leaving the network. Secondly, even if a static definition of self can be defined, the system may not be secure because a large number of attacks are perpetrated by trusted insiders rather than by outsiders. Finally, a large volume of network traffic from the Internet is potentially useful data, discarding all that data degrades the service quality of computer systems.
The first major impediment of currently proposed immune systems is the assumption that all data entering the network (or node) that is non-self is malicious. The second impediment to the creation of secure immune systems is the problem of computational complexity. The number of detectors that are required to match even a modest number
of features is very large and increases exponentially as the number of features increase. Forrest [21] use a 49-bit string composed of the source IP-address, destination IP-address and the destination port as the signature of the packets. To uniquely match all the possible 49-bit binary strings would require 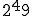 different detectors. Even ignoring half the bits would require
different detectors. Even ignoring half the bits would require 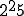 different detectors. This paper addresses the issue using two different approaches. The first approach is based on the premise that immune system needs to make informed choices so as to create fewer detectors with a higher probability of matching. The second approach is based on the premise that a more fundamental metric for classification of data be created, such as Kolmogorov Complexity.
different detectors. This paper addresses the issue using two different approaches. The first approach is based on the premise that immune system needs to make informed choices so as to create fewer detectors with a higher probability of matching. The second approach is based on the premise that a more fundamental metric for classification of data be created, such as Kolmogorov Complexity.
This paper also explores the concept of collective network defense to reduce computational burden on individual nodes. In this approach immune systems of different nodes work symbiotically and exchange pathogen information based upon their own detection schemes thereby assisting other nodes better select detectors to activate. The entire network can collectively pose a formidable barrier especially if the communication speed between the nodes is faster than the speed of proliferation of the pathogens. The two alternate schemes that can be explored are based on epidemiological models and random graphs. In Epidemiological models a central server collates all the information about virus detection at different nodes and then superposing this information on a spatial map the model predicates the rate and direction of the virus propagation. There are two problems with such a scheme, that is, speed and vulnerability of the central servers to attacks. Analysis and communication may take so long that the network may be overwhelmed. In the approach based on the theory of random graphs, messages can be propagated via hopping through chains of intermediaries.
A variant of the classical epidemiological model is a distributed model whereby a hierarchy of epidemiological servers exist; the server at the lowest level makes decision about the immediate neighborhood and sends the information up to the next level so that the information can be processed at higher levels for making global decisions such as shutting down, or throttling back, key routers and servers. This approach, though scalable and less vulnerable, requires significant overhead in managing flow of information. This approach is not investigated in this paper.
Work on understanding the basic concepts of the Immune System has proceeded for several years and primary concepts related to the function of the human immune system are well understood. However, work on developing Artificial Immune Systems is relatively new and many applications based on immunological concepts are emerging. Conceptual models of the immune system cover a broad spectrum of disciplines. So, for improved clarity, the extant literature is presented as four relevant streams of work: Biological Immune Systems, Artificial Immune Systems, Epidemiological Models, and Information Complexity.
Conceptual understanding of immune systems is based on several theories discussed in the literature including immune memory, clonal selection principle, idiotypic network theory, reinforcement leaning, and affinity maturation. Burnet [3] developed the clonal selection principle that states that each clone carries immunologically reactive sites corresponding to one or more antigens. Jerne [37] presented the idiotypic network theory that normal immune systems react against their own components to regulate themselves. Perelson and Oster [47] introduced the concept of general shape space that explains how a finite number of antibodies can detect an infinite number of antigens by clonal selection. Several methods have been proposed for analytic representation of the signatures of the pathogens that are matched in the immune system, such as bit-strings by Farmer [19], and De Boer and Perelson [16], Euclidean parameter spaces by Segal and Perelson [48], and polyhedron models by Weinand, [52].
Popularity of artificial immune systems is evident from the acceleration of research in its modeling over the last few years. Several applications based on immune systems outside the area of biological modeling have emerged recently, most notably computer security, which is a primary focus in this paper. Kephart [38][40] was perhaps the first to introduce the concept of using biologically inspired defenses against computer viruses and the use of immune systems for computer security. Forrest [23] also proposes the use of immune System concepts for design of computer security systems. Forrest et al. [21] provide an elaborate description of principles of immune systems that are applicable to computer security and present three alternate matching schemes: hamming distance, edit distance, and r-contiguous bits. They argue that the primary premise behind a computer immune system should be an ability to distinguish between self and non-self. They present a signature scheme where a data triple consisting of source IP-address, destination IP-address and the destination port number are used to identify self-packets from non-self packets. Hofmeyer [28][27] presents a detailed architecture of a computer immune system. He analytically compares different schemes for the detection of pathogens, such as hamming distance and specificity. There are several other works in the literature on the use of immune Systems for network security including those by Murray [45], Kim and Bentley [41], and Skormin [49].
Immune Systems are fairly complex with some unique schemes and are thus a rich source of concepts, which are applicable to a variety of problems. Hunt presents applications of computer immune systems in fraud detection and data mining. Ishida [32] presents an immune system for active noise control. Tarakanov and Dasgupta [51] present the architecture for a chip based on immunological paradigms. Tarakanov and Skormin [50] present a pattern recognition application using immuno-computing. Ishiguro et al. [33] present an application of immune systems in robotics for control applications. Dhasslear et al. [17] have used immune modeling for change detection algorithms. Hajela [26] presents applications of Immune Systems in Optimization.
Fred Cohen coined the word computer virus for self-replicating programs that spread on the network. Cohen and Murray suggest a link between computer viruses and biological epidemiology. Kephart [39] conducted the first comprehensive study of use of Epidemiology for studying computer viruses. They use a graph model for representing the network and study the virus propagation on the nodes of a graph using a differential equation based model. They deduct that no virus detection system is perfect, however, if the rate of detection is higher than the rate of propagation of viruses among nodes, the spread of viruses can be controlled. Williamson and Levine present a new epidemiological system dynamics model for virus propagation among network nodes. Their model consists of four states: susceptible, infected, detected, and removed; they provide a tool to perform simulations of virus propagation in networks. Some of the earlier work on epidemiology assumes homogeneity of the network. Moore [44] presents a model for disease propagation in a small-world graph. Barabási et al. [2] have shown that large networks are non-homogenous where high connectivity nodes are significant. Network topology can thus influence the spread of viruses. This research plans to address this issue by incorporating different network topologies in the model.
The complexity-based analysis technique presented in this paper takes into account the innovation of an attacker attempting to compromise an information system. A metric for innovation is not new, William of Occam suggested a technique 700 years ago [42]. The salient point of Occam's Razor and complexity-based vulnerability analysis is that the better one understands a phenomenon, the more concisely the phenomenon can be described. This is the essence of the goal of science: develop theories that require a minimal amount of information to be fully described. Ideally, all the knowledge required to describe a phenomenon can be algorithmically contained in formulae, and formulae that are larger than necessary lack a full understanding of the phenomenon. The ability of an attacker to understand, and thus successfully innovate a new attack against a system component, is directly related to the size of the minimal description of that component. It is in this sense that the proposed complexity-based signature detection algorithm has more "meaning" than simply detecting whether bits happen to match in various locations.
Consider an information system attacker as a scientist trying to learn more about his environment, that is, the target system. Parasitic computing [1] is a literal example of a scientist studying the operation of a communication network and utilizing its design to his advantage in an unintended manner. The attacker as scientist generates hypotheses and theorems. Theorems are the attacker's attempts to increase understanding of a system by assigning a cause to an event, rather than assuming all events are randomly generated. If theorem 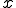 , described in bits, is of length
, described in bits, is of length 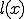 , then a theorem of length
, then a theorem of length 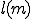 , where
, where 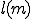 is much less than
is much less than 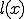 , is not only much more compact, but also
, is not only much more compact, but also 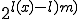 times more likely to be the actual cause than pure chance [42]. Thus, the more compactly a theorem can be stated, the more likely the attacker is to be able to determine the true underlying cause described by the theorem. Minimum description methods seek to estimate complexity by determining the smallest size to which a description can be compacted. Complex descriptions, containing a larger number of "random" interactions and lacking any form of repeatable patterns, cannot be described as compactly as simple descriptions. Kolmogorov Complexity is a complexity measure that falls within this category.
times more likely to be the actual cause than pure chance [42]. Thus, the more compactly a theorem can be stated, the more likely the attacker is to be able to determine the true underlying cause described by the theorem. Minimum description methods seek to estimate complexity by determining the smallest size to which a description can be compacted. Complex descriptions, containing a larger number of "random" interactions and lacking any form of repeatable patterns, cannot be described as compactly as simple descriptions. Kolmogorov Complexity is a complexity measure that falls within this category.
Thus, Kolmogorov Complexity is a measure of the descriptive complexity contained in an object. It refers to the minimum length of a program such that a universal computer can generate a specific sequence. A comprehensive introduction to Kolmogorov Complexity is contained in [43]. Kolmogorov Complexity is related to Shannon Entropy in that the expected value of K(x) for a random sequence is approximately the entropy of the source distribution for the process generating the sequence. However, Kolmogorov Complexity differs from entropy in that it relates to the specific string being considered rather than the source distribution.
A significant difficulty with Kolmogorov Complexity is that it is not, in general, computable. Any program that produces a given string is an upper bound on the Kolmogorov Complexity for this string, but one cannot compute the lower bound. Yet, as will be discussed later in this section, estimates have shown to be useful in providing information assurance and intrusion detection.
Kolmogorov Complexity is described in Equation 1, where 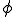 represents a universal computer,
represents a universal computer, 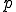 represents a program and
represents a program and 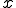 represents a string. Universal computers can be equated through programs of constant length, thus a mapping can be made between universal computers of different types. The string
represents a string. Universal computers can be equated through programs of constant length, thus a mapping can be made between universal computers of different types. The string 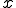 may be either data or the
description of a process in an actual system. Unless otherwise specified, consider
may be either data or the
description of a process in an actual system. Unless otherwise specified, consider 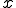 to be the program for a Turing Machine described in Equation 1.
to be the program for a Turing Machine described in Equation 1.
Kolmogorov Complexity has provided a useful framework from which to study objective metrics and methodologies for achieving information assurance. Recent results have shown promise for complexity estimators to detect FTP exploits and DDoS attacks [6][7][5]. Complexity is attractive as a metric for information assurance because it is an objective means of characterizing and modeling data and information processes for the purpose of benchmarking normal behavior, identifying weaknesses, and detecting deviations and anomalies.
This section describes the architecture of the immune system and the concept of collective defense. Section A describes the distributed security architecture based on the immune and epidemiology paradigms. Section B presents an analytic model of the proposed architecture.
In networked computer systems, network layer switches route information packets to nodes in the network. Traditionally, most virus detection software has been developed around single machines in the network, thus placing the entire burden of detection on individual machines. Conceptually, there is no reason why the computational burden of virus detection should not be distributed among the machines in the network as well as network components such as routers and switches. In fact, distributing the burden can enhance the resilience of networks to attacks. However, it demands an innovative model that is defensible on economic as well as technical grounds, because this model appears to differ from the old-fashioned network philosophy of keeping network components (intermediate nodes) as simple as possible while forcing complexity onto the edges (hosts) of the network. An innovative model is proposed by drawing analogies from the human immune system and epidemiology. This model's architecture has been designed to help answer the fundamental question of how much additional processing and capability should be added within the network as opposed to the edges of the network, particularly when additional capability is required to keep the network functioning when under attack. The model is described in two steps. Section A.1 describes the component on a single node while section A.2 describes the component distributed on the network.
When information packets arrive at switches or hosts, point of entry presents the first opportunity to inspect them for the presence of viruses. Since the packets have not been assembled into files at this stage, if we consider groups (or sequences) of packets, we can at best inspect them for existence of sectors of virus signatures. We will refer to this inspection as point-of-entry inspection. Because of the sheer traffic density and the size of the virus signature database, it becomes necessary to sample groups of packets for inspection at this first stage. That being the case, there is a possibility that viruses can escape detection at this inspection, either because the virus signatures may not be comprehensive, the virus signature straddles two contiguous sequences, or because the sequence containing the virus signature is not sampled. We call this the "false negatives" problem.
If the packets containing viruses escape detection during the above inspection, they are assembled into files that are carriers of the virus. When such files are executed, viruses attached to the carriers multiply and propagate. Therefore, there is a necessity to inspect all files for the existence of viruses. This second stage of inspection, which we will call carrier-level inspection, involves matching of the entire virus signature with the files either fully or by using indices of specificity as done in Forrest [22] Hofmeyer [27]. The mode of inspection (full inspection or sampling), and the frequency of inspection (real-time or periodic on files that have been changed) are decision parameters that can be chosen by network administrators.
Inspections are based on matching of virus signatures from a signatures database. At the point-of-entry inspection, matching is based on comparison of sectors of virus signatures with the group of packets sampled. Carrier-level inspections are based on comparison of the entire virus signature string with files stored on the node. In addition, because of the polymorphic and mutating nature of many viruses, it is necessary to compute the mutants of existing signatures in the database to be used at both levels of inspection. The Kolmogorov Complexity-based approach would quantify polymorphic distance from a given signature and thus would not explicitly require a complete set of polymorphic detectors.
At both levels of inspection, the number of detectors required will be very large if entire signatures or their sectors are used in matching. Therefore, as suggested by Forrest [23] we plan to implement matching based on a chosen specificity index, i.e., for a specificity index r, a match is said to exist if any r-contiguous bits in the signature (or its sub string) match with any r-contiguous bits on the sequence being inspected.
All of the above discussion applies to individual machines or components in the network. Each such entity is assumed to maintain its own signature database as well as immune system. The framework described above therefore provides reasonable assurance on the detection of viruses, but does not provide assurance on the prevention of the spread of such viruses. It is for the latter that we look to a metaphor from epidemiology. In addition, the previously discussed paradigm also introduces a large computational burden on the nodes. The epidemiological component of the model reduces this computational burden on individual network nodes as described in section A.2.
When a virus is detected by an individual entity on the network, it is important that information on such detections be maintained in a database so that their spread can be monitored. Such monitoring can be implemented either in a centralized fashion (as in many public health systems for the monitoring of the spread of diseases) or in a distributed fashion where each node in the network maintains an epidemiological database and monitors the spread of viruses in its neighborhood. Since any centralized monitoring facility is subject to denial-of-service attacks, in the model reported here, we introduce a distributed database of virus signatures and an epidemiological component to monitor the spread of viruses in the network neighborhood.
In the distributed model that we propose, when a node in the network detects a virus, the signature of such a virus is broadcast to all nodes in the immediate network neighborhood. Each node updates its own signatures database and performs statistical analysis of the virus detection frequencies. The results of such analysis, in conjunction with thresholds for responses can provide a resilient decision support system for dealing with viruses. The immune system and the epidemiological component form essential components of the network security infrastructure. The selection of detectors is based on stochastic models and each node ignores a large portion of the detector space. Collectively searching for viruses and sharing information allows a much larger search space to be covered by the immune system.
The average computational burden on each node would be minimized if each node were to compute a non-overlapping subset of non-self (malicious) space. In other words, the entire 'space' of non-self is partitioned into subsets, and each node computes a mutually exclusive part of the non-self detector set, thus distributing detector generation. It is only if/when a virus is detected by a corresponding detector of the non-self detector space (at a node responsible for that virus set) that the relevant detector set is broadcast to other vulnerable nodes in the network. In this case both processing and memory overhead can be reduced since each node handles only a subset of the entire non-self space at the cost of quickly moving non-self information to the correct location when viruses are imminent. However, the initial partitioning of non-self spatially across the network becomes a critical design issue. There is management overhead in initiating and maintaining such a partitioning.
If a virus is polymorphic and Kolmogorov complexity proves to be sufficient in matching an evolutionary chain of such viruses, then Kolmogorov Complexity would be a key component in properly locating the correct set of detectors in proper locations. A self-organized system in which detectors would migrate towards regions where discrepant complexity patterns are observed could address the maintenance issues associated with maintenance of detectors.
An obvious problem with the approach is the amount of damage that would be sustained, in the worse case, if a virus entered and spread throughout the network before reaching the spatial location in which the corresponding detectors were generated. In this case, one would almost wish to promote the spread of the virus in order to guarantee that the portion of the network with the corresponding detector set is reached quickly.
A subset of the space, however, does not necessarily have to be exclusive to a node. It has been shown in the literature that in random graphs the average degree of connectivity of each node is very small. Recent studies have also indicated that many networks, including the world wide web follow a scale-free degree distribution in which the fraction of sites having k connections follows a power law: P (k) = ck. The average distance between sites in scale-free networks is much smaller than that in regular random networks. If instead of a mutually exclusive set of detectors at each node, an overlapping set of detectors was created across network nodes, even with a modest connectivity of the graph, the virus should be able to reach a node with the specific detector quickly. An optimization model can be created to partition and replicate the non-self space so as to have an acceptable response rate. According to Kephart and White [39], as long as the rate of detection and cleanup is higher than the infection rate, the system will not get overwhelmed.
The alternative to the approach described above is to let each node randomly select detectors from the detector set. This approach has low maintenance overhead however, it may leave a significant portion of the search space unexplored based on the number of nodes on the network, the specificity used in detection and the sampling rate at each node. The paper explores the feasibility of using each of these approaches via an analytic model.
An analytic model was developed to investigate the behavior of the immune system in response to entry of a pathogen into the network. The model considers the entry and propagation of a single pathogen in the network. Since the arrival or generation of any virus strain is independent of any other virus strain in the network, the model can be extrapolated for multiple viruses entering the network without loss of generality. A detector, 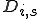 , is defined with identifier
, is defined with identifier 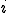 and specificity
and specificity  . The symbol
. The symbol 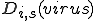 represents the relation of a specific detector
represents the relation of a specific detector 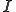 to a specific signature,
to a specific signature, 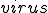 . A single detector can match many sequences, however, this notation refers to relationships between a specific detector and a specific potential virus signature, such as evolving a detector to become more specific to a virus signature. A detector match to a specific virus is represented by
. A single detector can match many sequences, however, this notation refers to relationships between a specific detector and a specific potential virus signature, such as evolving a detector to become more specific to a virus signature. A detector match to a specific virus is represented by 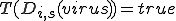 . The model assumes that self-space is completely known, however, holes may exist in the non-self space. The initial set of detectors covers as much of nonself as possible with low specificity, that is,
. The model assumes that self-space is completely known, however, holes may exist in the non-self space. The initial set of detectors covers as much of nonself as possible with low specificity, that is, 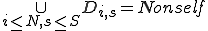 , where
, where 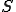 is a given specificity and
is a given specificity and 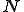 is the number of detectors required to cover nonself at specificity
is the number of detectors required to cover nonself at specificity 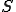 . The probability of a hole
. The probability of a hole 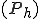 is greater at lower specificity. Thus, we can only approximate coverage of nonself space with low specificity. Holes are created when detectors that match self-signatures are eliminated from the non-self detector set during clonal deletion. However, by using low specificity coverage of nonself is achieved more quickly and with fewer detectors. The non-self detectors are distributed across the network such that multiple detectors of the same type can exist on the network, however, any node on the network can have only one copy of a specific detector. Multiple copies of detectors
is greater at lower specificity. Thus, we can only approximate coverage of nonself space with low specificity. Holes are created when detectors that match self-signatures are eliminated from the non-self detector set during clonal deletion. However, by using low specificity coverage of nonself is achieved more quickly and with fewer detectors. The non-self detectors are distributed across the network such that multiple detectors of the same type can exist on the network, however, any node on the network can have only one copy of a specific detector. Multiple copies of detectors  are distributed on nodes throughout the network, thus for any given node
are distributed on nodes throughout the network, thus for any given node 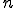 ,
, 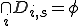 , that is, a node may contain multiple low specificity detectors only if they are of different types. An optimization algorithm is used to determine the initial distribution as well as redistribution of detectors in the network. The goal of the optimization is to minimize the damage potential of pathogens by redistributing the detectors on the network in response to changes in network configuration, network latency and perceived threats.
, that is, a node may contain multiple low specificity detectors only if they are of different types. An optimization algorithm is used to determine the initial distribution as well as redistribution of detectors in the network. The goal of the optimization is to minimize the damage potential of pathogens by redistributing the detectors on the network in response to changes in network configuration, network latency and perceived threats.
There are two kinds of detectors that cover the non-self space: low-specificity and high specificity. The low specificity detectors, the original 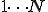 detectors, cover the bulk of non-self space and high specificity detectors correspond to existing known viruses. The distribution of low specificity detectors is coordinated centrally, however
each network node maintains its own set of high specificity detectors. The purpose of the high-specificity detectors is to present a targeted response to known pathogens and to patch the holes that arise in the detector space. Creation and distribution of high specificity detectors is computationally expensive since a relatively large number of detectors are required as compared to low specificity detectors to cover the same detector space. Lower the specificity of detectors the less detectors are required however the lower the specificity greater the chances of holes in the detector space requiring more specific detectors. Thus the specificity of the detectors needs to be optimized to minimize the detector count.
detectors, cover the bulk of non-self space and high specificity detectors correspond to existing known viruses. The distribution of low specificity detectors is coordinated centrally, however
each network node maintains its own set of high specificity detectors. The purpose of the high-specificity detectors is to present a targeted response to known pathogens and to patch the holes that arise in the detector space. Creation and distribution of high specificity detectors is computationally expensive since a relatively large number of detectors are required as compared to low specificity detectors to cover the same detector space. Lower the specificity of detectors the less detectors are required however the lower the specificity greater the chances of holes in the detector space requiring more specific detectors. Thus the specificity of the detectors needs to be optimized to minimize the detector count.
The immune system engages in a process called affinity maturation that results in the creation of specific detectors in response to detection of pathogens by low specificity detectors. When a non-self packet is detected on a node the immune system rapidly generates copies of the detector that matched it by cloning the detector and using a high mutation rate to generate diversity in the population. The generated clones themselves undergo the process of cloning and mutation to generate more detectors. The rate of cloning is proportional to the fitness of the detectors and is defined as the detector specificity to the observed pathogen. The fitness function 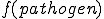 assigns higher fitness to detectors such that specificity for a suspected pathogen is increased, such that,
assigns higher fitness to detectors such that specificity for a suspected pathogen is increased, such that, 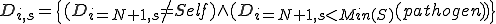 that is, a new detector is created with identifier
that is, a new detector is created with identifier 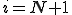 , and specificity with regard to the suspected pathogen is lower,
, and specificity with regard to the suspected pathogen is lower, 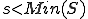 where
where 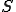 is the set of specificity from detectors such that
is the set of specificity from detectors such that 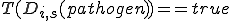 . The low specificity detectors
. The low specificity detectors 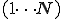 remain resident as originally distributed upon each node. The higher specificity detectors
remain resident as originally distributed upon each node. The higher specificity detectors 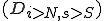 are created in response to the appearance of a virus and migrate from node to node. Evolution towards better fitness as defined by
are created in response to the appearance of a virus and migrate from node to node. Evolution towards better fitness as defined by 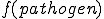 is induced by two mechanisms: (1) at least one of the low specificity nonself detectors is activated (2) indication of damage. A damage model provides a framework in which the occurrence of damage to node
is induced by two mechanisms: (1) at least one of the low specificity nonself detectors is activated (2) indication of damage. A damage model provides a framework in which the occurrence of damage to node 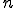 can be assessed based upon for example, excessive load, excessive, rapidly escalating connectivity, loss of files, etc... Damage will be defined in this paper as an abstract value
can be assessed based upon for example, excessive load, excessive, rapidly escalating connectivity, loss of files, etc... Damage will be defined in this paper as an abstract value 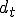 , which represents the damage at a given time such that
, which represents the damage at a given time such that 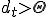 indicates that damage has exceeded a threshold,
indicates that damage has exceeded a threshold, 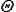 . The trigger is defined by a Boolean function
. The trigger is defined by a Boolean function 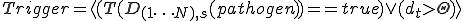 . Once triggered by damage detection, an
evolutionary search occurs to find a specificity that more tightly matches the pathogen based upon either the low specificity detector, that is, detector identifiers
. Once triggered by damage detection, an
evolutionary search occurs to find a specificity that more tightly matches the pathogen based upon either the low specificity detector, that is, detector identifiers 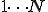 , or if the pathogen lies in a hole in the detector space fill holes that overlap with the most recent changes to self. Any mutated clones that are not exclusive of self are destroyed. The fittest clone is then distributed to the other nodes of the network to assist in the identification and destruction of the pathogen. In the scenario when the signature of the packet is neither identified as self nor as non-self co-stimulation from multiple nodes is used to designate a packet as benign or malicious. Co-stimulation can be based on damage detection on the network as well as on the proximity of the signature to specific detectors on multiple network nodes.
, or if the pathogen lies in a hole in the detector space fill holes that overlap with the most recent changes to self. Any mutated clones that are not exclusive of self are destroyed. The fittest clone is then distributed to the other nodes of the network to assist in the identification and destruction of the pathogen. In the scenario when the signature of the packet is neither identified as self nor as non-self co-stimulation from multiple nodes is used to designate a packet as benign or malicious. Co-stimulation can be based on damage detection on the network as well as on the proximity of the signature to specific detectors on multiple network nodes.
The stochastic model for the immune system is shown in Figure 1. It shows the behavior of a node in response to insertion of a specific pathogen into the network. Each node can have one of three states, that is, susceptible (S), immune (I) or infected (D). A node is in immune state if it contains a detector for a specific pathogen. It is in susceptible state if it neither contains the pathogen nor the detector and it is in infected state if it contains the pathogen but not the detector. The possible state transitions in the model are listed below and the initial state vector is presented along with the specific plots of the simulation.
We model the network as a discrete-time stochastic process to determine how the state of the system changes at discrete points in time. A generic discrete-time stochastic model is first presented and then a specific model representing the immune network is presented. Let S denote the states of a node.
![\mbox{S = [S_1, S_2, \dots, S_n]}](../img/cache/a1c528b20f2600d8df8c41d1fd8747d3.gif)
Where n is the total number of states, Let 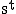 be the probability distribution of the states at time t.
be the probability distribution of the states at time t.
![\mbox{s^t = [s_0, s_1, \dots, s_n] such that} \sum_{j = 1}^{j = n}{}s_j = 1 \text{ and }0 \le s_i \le 1.](../img/cache/89099fe431e7ab3987a3995581ded457.gif)
Let T be the transition matrix where 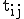 is the probability of transition from state i to state j such that
is the probability of transition from state i to state j such that 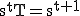
![T = \left[\begin{matrix}t_{11}&t_{12}&\dots&t_{1n}\\t_{21}&t_{22}&\dots&t_{2n}\\\vdots&\vdots&\vdots&\vdots\\t_{n1}&t_{n2}&\dots&t_{nn}\end{matrix}\right]](../img/cache/1db883284a2d7b2874d1dfa651f1dd30.gif)
Here, 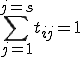
Probability that a sequence of states,  at time 1, 2, ... k is then given by
at time 1, 2, ... k is then given by
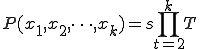
The probabilities of transition of an individual node depends on the network topology parameters such as the node connectivity and the distribution of detectors on the network, as well as the rate of execution of programs on a machine. Let us consider the case where no detectors are present on the network. From the state transition diagram in Figure 1, S=[s, i, d].
![T = \left[\begin{matrix}t_{ss}&t_{si}&t_{sd}\\t_{is}&t_{ii}&t_{id}\\t_{ds}&t_{di}&t_{dd}\end{matrix}\right]](../img/cache/938c6b25362bea75dd4caf04991947e5.gif)
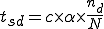
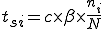
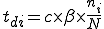
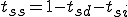
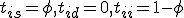
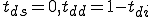
From Figure 17, we can estimate the rate of detector generation for a node. From Figure 12 and Figure 16, we know the probability of match of a detector for a given specificity and sequence length. These can be used to help determine 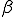 in the stochastic model.
in the stochastic model. 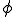 in the stochastic model represents the potential for a polymorphic virus to change its signature and thus evade detection. The steep slopes in Figure 12 and Figure 16 indicate that the rate of change of match probability
in the stochastic model represents the potential for a polymorphic virus to change its signature and thus evade detection. The steep slopes in Figure 12 and Figure 16 indicate that the rate of change of match probability 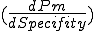 is large and thus very sensitive to specificity. A small change in signature causes a drastic change in the detector's ability to match the virus, thus increasing the number of detectors required and perhaps impacting
is large and thus very sensitive to specificity. A small change in signature causes a drastic change in the detector's ability to match the virus, thus increasing the number of detectors required and perhaps impacting 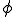 . Several questions can be answered using the stochastic model. The model uses as assessment metric of damage as
. Several questions can be answered using the stochastic model. The model uses as assessment metric of damage as 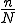 nodes infected and bases the time to infect on the hop count.
nodes infected and bases the time to infect on the hop count.
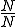 nodes infected, given the virus propagation rates and detector locations from (4) above?
nodes infected, given the virus propagation rates and detector locations from (4) above?The research addresses questions 1 & 2 described above with questions 3-7 deferred for future work. In the simulations presented here the stochastic model is evaluated for the case of 100 nodes. Stochastic parameter values are shown at the top of each graph. In Figure 2 there is a relatively high rate of detector release (0.8) into the network. The plots show that number of immune nodes grows rapidly. In Figure 3, detector release rate drops to 0.4 resulting in a slower and lower immune rate among the nodes. In Figure 4 the detector release rate has further dropped to 0.2 and the virus infection rate climbs up to 0.8. This results in a dangerous situation in which the number of immune nodes gradually decreases while the number of infected nodes increases. The generation and release rate of detectors is critical, given any rate of virus propagation. Figure 17 later in the paper shows the rate of detector generation as a function of size of signature. Even in the best case when the self is small and the ability to generate nonself is small the rate of detector generation is very low showing that a system of realistic size would take an unreasonable long time to cover all of nonself. (assuming random detector generation and deleting detectors that match self).
The impact of network connectivity on the stochastic model can be seen in the network topologies shown in Figure 5. The stochastic model was applied to these topologies. In Figure 6, the stochastic model is applied to network Figure 5.a that has relatively low connectivity, the probability of connection between any two nodes is 0.3 and the network diameter is 4. The stochastic model parameters in the figures in this paper are set to model a faster detector generation and propagation rate than virus propagation rate, thus the detectors take advantage of network connectivity spread faster than the virus. Greater network connectivity increases the rate of spread of both detectors and viruses, with a more rapid spread of detectors, causing the number of immune nodes to increase at a more rapid rate than in a low connectivity network. Notice that with this low level of connectivity the point where the probabilities of susceptible and immune cross one another is beyond the right side of the visible graph. In Figure 7, the stochastic model is applied to the network in Figure 5.b, which is slightly more connected, with a probability of connection is 0.5 and network diameter is 3. Here the crossover point from susceptible to immune occurs around transition 28. Finally, in Figure 8, the stochastic model is applied to the network shown in Figure 5.c, which is more fully connected, with a connection probability between any two nodes of 0.8 and a diameter of 2. Here the crossover point from susceptible to immune occurs even earlier, near transition 17. The work on modeling of the virus spread and detector propagation is preliminary. Figure 9 shows the results from an immune system simulation, which shows that specificity is that the detection rate is strongly dependent on the specificity. At high specificity it is not possible to have a large enough sampling rate to efficiently run the immune system. The authors intend to use the existing results and extend their modeling further to answer some of the remaining questions presented above.
A critical objective of a signature-based virus detection system is to generate and match detectors with potentially transient software components, e.g. packets, drivers, patches, etc... such that the frequency of matches to legitimate system components is minimized (preferably zero) while the matching frequency of illegal components is maximized (preferably all). There is considerable overhead and time required to generate all detectors that do not match a given system, referred to as "self". However, design tradeoffs must be made among several dimensions when optimizing the efficiency of matching detectors with potentially illegal component sequences, or signatures. Contiguous element matching, number of total matches, and a novel Kolmogorov Complexity-based matching technique are compared and contrasted in terms of their matching efficacy this paper.
Among many parameters affecting detector overhead are sequence length and generation time of each detector; some matching techniques attempt to minimize this overhead by allowing one signature to match many possible nonself-sequences. This is accomplished by allowing wildcard positions in the detector that can match any element. In a similar manner, a `degree' of match can be specified by such concepts as contiguous element matching or number of total matches. Allowing one detector to match more than one component in a system, while reducing overhead in generating detectors by allowing fewer detectors to `cover' a larger space, introduces another problem, namely, holes. Holes are virus signatures that cannot be matched by detectors because legitimate system components would be matched by the same detector. In this paper we will refer to matching techniques that allow detectors to match many potential components as `loose' matching techniques. Those techniques that allow only a one-to-one or one-to-few detector - to-sequence matches will be referred to as `tight'. Tightness and looseness of matching are one of the tradeoffs involved in determining optimal detector performance. Three matching techniques are analyzed in this paper, namely, contiguous element matching, number of total matches, and a degree of match based upon estimates of Kolmogorov Complexity. The term "complexity" will be used in the remainder of this paper in reference to estimates of Kolmogorov Complexity. A comparison and contrast between specificity and Kolmogorov Complexity is illustrated in Figure 10. Specificity is illustrated on the left side if the figure. Detectors with a small or tight specificity can exactly cover the nonself sequences illustrated in the figure. However, large or loose specificity requires fewer detectors but can create holes. A Kolmogorov Complexity-based distance metric illustrated on the right side of Figure 10. The complexity-based metric examines differences in complexity between self and nonself sequences as well as measuring similarity among nonself sequences. The symbols used in the analysis are shown in Table 2. The technique of generating all potential detectors and eliminating those that match legal components is assumed.
Contiguous element matching returns the length of the longest set of contiguously matching elements between a detector and a target sequence as illustrated in Figure 13. The detector is the middle sequence and the target strings above and below the detector. The detector and target sequence are the same length. The target sequence along the top in non-sliding, in other words, the sequences are lined along side one another and the longest contiguous set of matching elements is counted. In Figure 13, the longest set of contiguously matching elements is has three elements as shown within the leftmost dashed box. Equation 3, derived in [46], shows the equation used to obtain the probability of match assuming that signatures are randomly generated. The probability of match assumes a randomly generated string with m possibilities for each element. Each position can match with probability 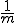 and does not match with probability
and does not match with probability 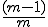 . There is an
. There is an 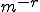 chance that
chance that  contiguous elements will match and there are
contiguous elements will match and there are 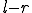 chances for a run of length
chances for a run of length  in a sequence of length
in a sequence of length 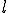 . Each run is preceded by a mismatch with probability
. Each run is preceded by a mismatch with probability 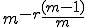 . A plot of the RePast simulation and contiguous element matching results are shown in Figure 12. Another form of contiguous element matching allows for one sequence to slide alongside another while searching for the longest contiguous sequence of matching elements. This is illustrated in the target sequence at the bottom of Figure 13. Each time the target sequence is shifted one character at a time to the right, the longest contiguous match is determined. After the target sequence has been shifted completely across the detector, the longest set of longest
contiguous matches, in this case four as shown in the right-most dashed box in Figure 13, is defined as the sliding specificity.
. A plot of the RePast simulation and contiguous element matching results are shown in Figure 12. Another form of contiguous element matching allows for one sequence to slide alongside another while searching for the longest contiguous sequence of matching elements. This is illustrated in the target sequence at the bottom of Figure 13. Each time the target sequence is shifted one character at a time to the right, the longest contiguous match is determined. After the target sequence has been shifted completely across the detector, the longest set of longest
contiguous matches, in this case four as shown in the right-most dashed box in Figure 13, is defined as the sliding specificity.
The number of total matches is quantified by simply counting the total number of elements that match between a detector and target sequence, regardless of whether the matches are contiguous. This is illustrated in Figure 14. The dashed boxes show the matches. The equation for total match frequency can be derived most simply from the previous derivation (for contiguous matches) by noting that the probabilities of match 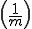 and non-match
and non-match 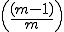 are simply summed as a binomial coefficient and multiplied by the opportunity for the signature to slide alongside the sequence under test as shown in Equation 4.
are simply summed as a binomial coefficient and multiplied by the opportunity for the signature to slide alongside the sequence under test as shown in Equation 4.
A sliding total match count allows one sequence to slide alongside another, while the largest set of matching elements is determined after each right shift of the target string. In both the contiguous and total number of matching elements techniques, allowing the target sequence to slide allows for more opportunities for matches to occur.
A Kolmogorov Complexity-based approach to signature matching is motivated by the fact that there is little `meaning' behind quantitative measures of bit-matching techniques, such as contiguous element matching, and the number of total matches. A brief review of Kolmogorov Complexity recently applied to communications can be found in areas such as Active Networks [9], Network Management [4], and Information Assurance [7] [5]. One of the most complete references on the subject can be found in [43] and an introductory discussion of complexity and meaning can be found in [42].
Complexity is defined as the smallest program that can generate the natural number that describes the binary sequence 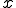 . It is the smallest program, or algorithm, that describes sequence
. It is the smallest program, or algorithm, that describes sequence 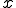 . This definition states that a bit-string,
. This definition states that a bit-string, 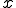 , that requires a larger smallest algorithm to describe the bit-string is quantifiably more complex. Consider a very basic definition of complexity,
, that requires a larger smallest algorithm to describe the bit-string is quantifiably more complex. Consider a very basic definition of complexity, 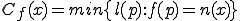 , where
, where 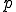 is a Turing Program,
is a Turing Program, 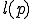 is the length of program
is the length of program 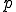 , and
, and 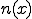 comes from [43] in which an unambiguous mapping is made from binary values to positive
integers. Complexity is thus defined as the smallest program that can generate the natural number that describes the binary sequence
comes from [43] in which an unambiguous mapping is made from binary values to positive
integers. Complexity is thus defined as the smallest program that can generate the natural number that describes the binary sequence 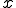 .
.
In general, a wide range of programs, or algorithmic descriptions can generate a bit-string. Often, it is the case that the size of a program, or with regard to the definition above - the algorithmic description, can be traded-off for speed. Thus, we need to be careful to distinguish the above definition of complexity from space and time complexity. From the standpoint of bandwidth, we desire small size, namely, the complexity, which must be approximately equal to, or smaller than, the static data whose complexity is being computed. However, from the standpoint of processing speed, faster, but possibly larger code may be desired.
Consider, 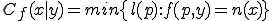 . In this case, the complexity of
. In this case, the complexity of  is conditioned upon
is conditioned upon  . One can think of this as meaning that executing program
. One can think of this as meaning that executing program 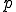 with input
with input 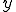 generates
generates 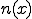 . Conditional complexity is important because an active packet can contain both code and data, the proportion of which may vary. Consider
. Conditional complexity is important because an active packet can contain both code and data, the proportion of which may vary. Consider 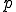 as the code in an active packet,
as the code in an active packet, 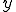 may be a conditional bit-string for determining conditional complexity as static data in the packet, and
may be a conditional bit-string for determining conditional complexity as static data in the packet, and 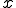 is the final "piece" of information to be sent, represented as a bit-string. The function
is the final "piece" of information to be sent, represented as a bit-string. The function 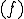 is the code within a network processor (NodeOS/EE) [9] [10] upon which the packet is executed.
is the code within a network processor (NodeOS/EE) [9] [10] upon which the packet is executed.
Note that any protocol 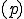 carried in the active packet can have its complexity estimated in this manner. Thus, complexity as an information assurance measure is already obtained as part of complexity estimation. Complexity also plays a role in evolution, and could well play a role in technical advances, namely robust evolutionary or genetic programming within a running network for protocol generation and support. With regard to signature matching, one can very roughly summarize a Kolmogorov Complexity-based degree of match by the sketch shown in Figure 15; it is the length, in bits, of the smallest Universal Turing Machine Program that can convert a sequence under test to a given signature. A matching signature that matches a detector would require a program of length zero, while a sequence that does not match in any element positions would require a program at least as long as a description of the detector itself.
carried in the active packet can have its complexity estimated in this manner. Thus, complexity as an information assurance measure is already obtained as part of complexity estimation. Complexity also plays a role in evolution, and could well play a role in technical advances, namely robust evolutionary or genetic programming within a running network for protocol generation and support. With regard to signature matching, one can very roughly summarize a Kolmogorov Complexity-based degree of match by the sketch shown in Figure 15; it is the length, in bits, of the smallest Universal Turing Machine Program that can convert a sequence under test to a given signature. A matching signature that matches a detector would require a program of length zero, while a sequence that does not match in any element positions would require a program at least as long as a description of the detector itself.
The technique to estimate Kolmogorov Complexity used in this work is based upon a complexity differential, illustrated in Figure 11. Sequences are partitioned into fixed length packets. The sum of the inverse compression ratio of the individual packets is compared to the fully concatenated sequence, that is, a single block consisting of the entire sequence. A complexity differential is defined as the difference between the cumulative complexities of individual packets and the total complexity computed when those packets are concatenated to form a single packet. If
packets 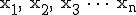 have complexities
have complexities 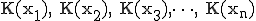 , then the complexity differential is computed as:
, then the complexity differential is computed as: ![\mbox{[K(x_1) + K(x_2) + K(x_3) +\dots+ K(x_n)] - K(x_1x_2x_3\dots x_n)}](../img/cache/fe30932b94372ea80944d4393741f36d.gif) , where
, where 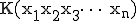 is the complexity of the packets concatenated together. If packets
is the complexity of the packets concatenated together. If packets 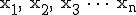 are completely random,
are completely random, 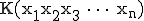 will be equal to the sum of the individual complexities and the complexity differential will therefore be zero. However, if the packets are highly correlated i.e. some pattern emerges in their concatenation, then the concatenated packet can be represented by a smaller program and hence its complexity i.e.,
will be equal to the sum of the individual complexities and the complexity differential will therefore be zero. However, if the packets are highly correlated i.e. some pattern emerges in their concatenation, then the concatenated packet can be represented by a smaller program and hence its complexity i.e., 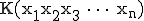 will be smaller than the cumulative complexity. The complexity differential is the estimate used in the experiments in this paper.
will be smaller than the cumulative complexity. The complexity differential is the estimate used in the experiments in this paper.
This section compares and contrasts results regarding the performance of the signature matching techniques defined in the previous section. Results from a RePast simulation are compared with analytically derived results from a Mathematica immunological package under development as part of this research. To the right of Figure 12, analytical and RePast model results are shown for the matching probability as a function of detector length and number of contiguous elements required, that is, the specificity. However, it is suggested later in this paper that specificity is not as useful a measure of similarity as Kolmogorov Complexity estimates.
To the left of Figure 12, more detail regarding simulation versus analytical results for one slice of the surface shown in the previous figures. The analysis and the simulation match very nicely. The important point of the surfaces is that the probability of match drops off precipitously as either sequence length decreases or the number of contiguous matches required (specificity) increases. Contiguous matching results in a very brittle matching probability.
The number of total matching elements analytical and simulation results is shown to the left and right respectively of Figure 16. Analytical and simulation results appear to match reasonably well. The matching probability, although greater than contiguous element matching as expected, again drops off suddenly with decreasing sequence lengths or increasing specificity. This implies that both contiguous matching and number of total elements matched provide very little control over the degree of matching. Later in this section we consider a complexity-based approach that provides a finer control over degree of matching as well as more potential meaning to the degree of match.
The overhead of generating detectors is illustrated in Figure 17. The rate of detector generation is based upon a Java RePast implementation in which random detector sequences were generated at a measured expected rate of approximately one 4000-element sequence per millisecond. Non-sliding contiguous matches among these sequences was measured to have an expected execution time of approximately eight milliseconds. Measured results for the other matching technique are shown in Figure 18, namely the complexity estimation techniques based upon Zip (Zip), Limpel-Zev (LZ), and Entropy, which will be discussed in the next section. If detectors match the legitimate system (self) then they are eliminated, resulting in a zero detector production rate. When detectors do not match self, they require the time that it takes to attempt to match all self sequences. Thus, total detector generation time is the time to create a random detector sequence plus the expected time to match all self-sequences. It is apparent from Figure 17 that detector generation rate drops dramatically with the size of self. Thus sequence-matching time is a significant factor in the ability of this type of immune response to react in a timely, or even feasible, manner. In the RePast implementation, it is apparent that the differential complexity estimation techniques based upon Entropy and Zip are on the same order of performance as simple contiguous and total matching.
An experiment designed to compare and contrast complexity estimation versus the previously discussed signature matching techniques was comprised of detecting whether there is a correlation between traditional techniques and complexity estimation. In order to obtain sequences that are nearly realistic, the Anna Kournikova virus code was used as input for our correlation analysis. We began with a sequence containing the entire virus definition, and then selected a chunk of characters randomly from the sequence. The chunks were concatenated until a 4000-character sequence was assembled. A set of 1000 such virus target sequences was generated. A single randomly generated 4000-character detector was also generated. We matched the virus sequences with the detector using all the matching techniques described in this paper in order to obtain various forms of specificity, namely: contiguous matches, sliding contiguous matches, total matches, sliding total matches, and complexity estimations, namely: entropy, Zip, and LZ. We wished to examine what degree of correlation exists among the various forms of specificity as well as between specificity and complexity.
Table 3 shows statistical correlation results among traditional signature detection techniques for the 4000-character sequences. It is apparent that both sliding techniques (highlighted in yellow) show relatively high correlation with each other (0.52) while both non-sliding techniques (highlighted in blue) show relatively high correlation (0.56). The sliding and non-sliding techniques show relatively low correlation with one another.
Table 4 shows correlation among the differential complexity estimators (Entropy (H), Zip, and Limpel-Zev (LZ)) and the traditional signature matching techniques. In this table there is a relatively high correlation among the Zip differential complexity estimator and both sliding techniques. The reason for the higher correlation between complexity and sliding techniques is that complexity estimation identifies patterns throughout the entire length of the detector and target sequences being matched, rather than searching for one instance of a longest match.
It is hypothesized that a mutating virus would easily escape detection by traditional matching mechanisms due to the brittle matching probability illustrated in the surface plots of Figure 12 and Figure 16. Degree of match would be more readily apparent and meaningful using a complexity-based technique. In fact relationships between positions of matching bits and evolutionary changes in program code, such as modifications to a virus either by human intervention or via automated polymorphic viruses would be missed by current techniques while being measurable via complexity-based techniques.
We further hypothesize that signatures may be eliminated if one can distinguish self from nonself on the bases of complexity estimates alone, thus eliminating the need to generate and test large numbers of detectors. This technique also removes the burden of memory required to hold and maintain large sets of detectors. The challenge will be to determine the discriminatory capability versus overhead of complexity estimation versus traditional matching mechanisms.
This paper presents a paradigm for a network security that integrates defenses at the node and the network level. In this paradigm, that we term as collective defense, complexity is used a function to integrate the biological paradigms of epidemiology and immunology. In this model the burden of security is shared by distributing the detectors among network nodes. The detectors can replicate and migrate across the networks based on the concentrations of pathogens in different parts of the network. A stochastic model has been developed to analyze the impact of detector versus virus propagation rates in such a system. We have also developed a simulation based on Repast that models this collective defense framework. Some preliminary results show a strong relationship between specificity and complexity as well as a correlation between estimates of complexity and various measures of specificity. We've hypothesized that complexity estimation may be a better matching mechanism for virus signatures, particularly for relating polymorphic viruses because Kolmogorov Complexity estimation attempts to account for model size used to classify information and provides a measure of the fitness of an explanation used in classification. The preliminary results compare and contrast the efficiency of different virus signature detection schemes. It is hypothesized that Information Distance, that is based on Kolmogorov Complexity will provide a reasonable measure of similarity for a virus signature. In particular, Information Distance will work significantly better for polymorphic viruses because such viruses would be most likely to evade detection and survive by using a small mutation algorithm, namely, one that approaches the conditional Kolmogorov Complexity estimate between the original virus string and the new virus string.
The authors would like to thank Professor Jagdish Gangolly (Department of Accounting & Law, University at Albany, SUNY) for his helpful comments and review of the manuscript.
Equation 1: Definition of Kolmogorov Complexity.
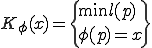
Equation 2: Conditional Kolmogorov Complexity.
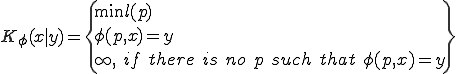
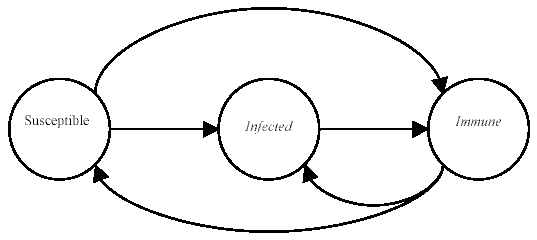
Figure 1: State Transition Diagram
Table 1: Symbols Used in the Stochastic Model.
| Symbol | Explanation |
|---|---|
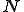 | Number of Nodes in the Network |
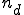 | Number of dirty nodes (infected) |
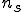 | Number of susceptible nodes (non-infected) |
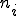 | Number of nodes that are immune |
 | Average number of connections of each node |
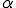 | Probability virus release from infected node |
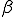 | Probability of detector release from immune node |
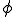 | Rate of transition from Immune back to Susceptible |
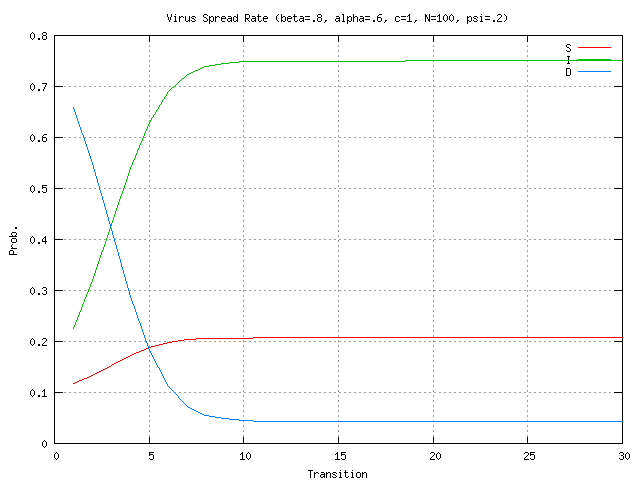
Figure 2: High Detector Generation/Release Rate.
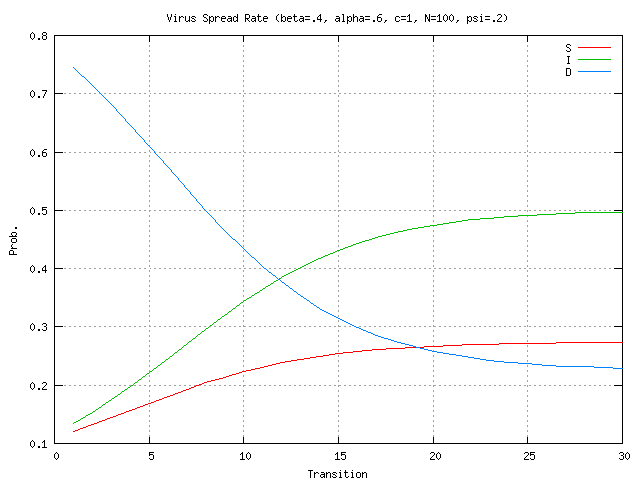
Figure 3: Lower Detector Generation/Release Rate.
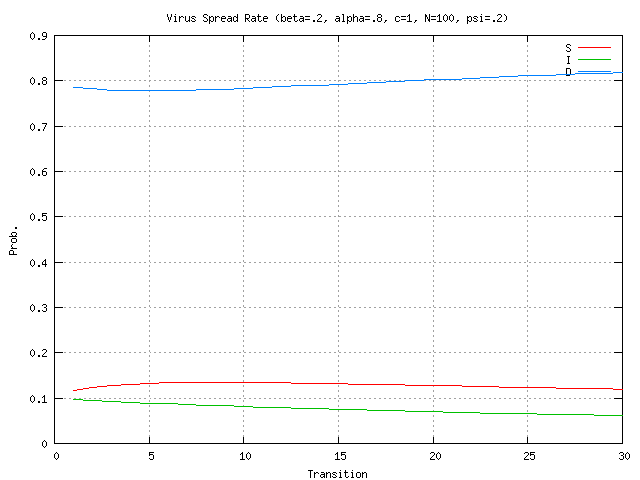
Figure 4: Very Low Detector Generation/Release Rate.
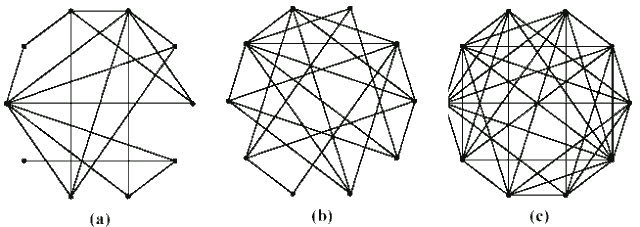
Figure 5: Random Graphs (a) Probability of Connection 0.3 Diameter 4 (b) Probability of Connection 0.5 Diameter 3 (c) Probability of Connection 0.8 Diameter 2.
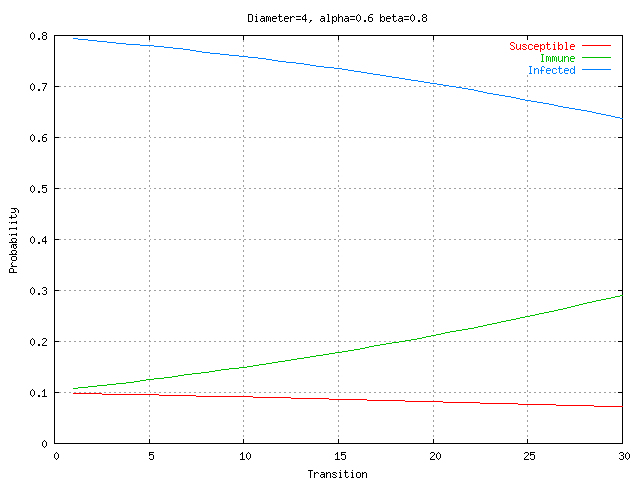
Figure 6: Impact of Large Diameter, Low Connectivity Graph on Stochastic Model.
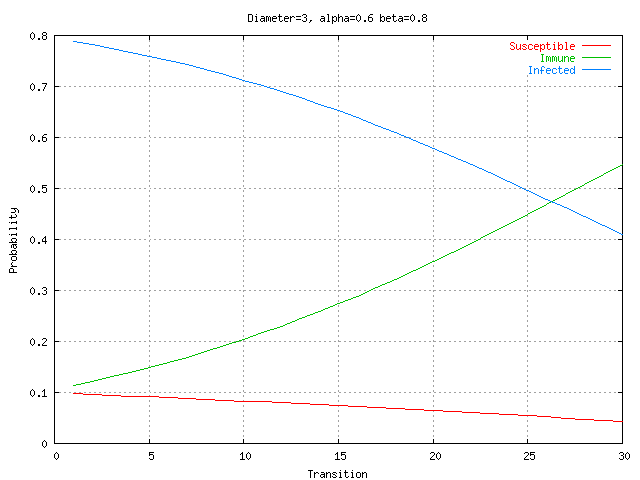
Figure 7: Impact of Medium Diameter Graph on Stochastic Model.
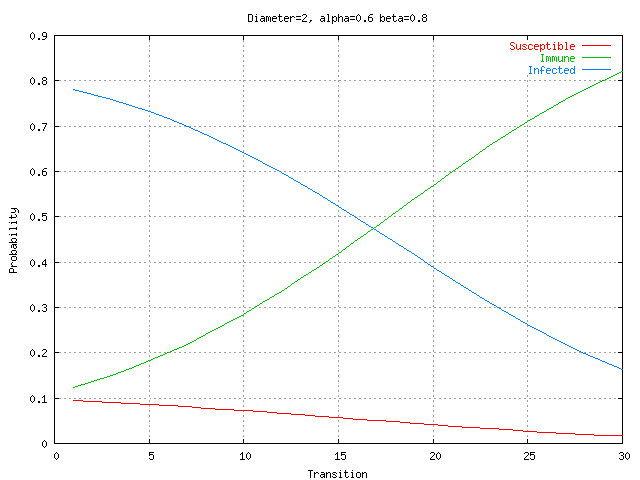
Figure 8: Impact of Low Diameter, High Connectivity Graph on Stochastic Model.
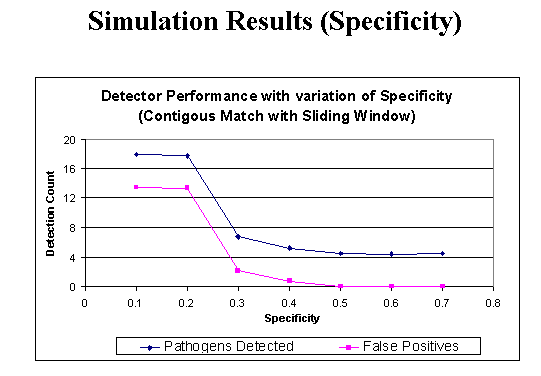
Figure 9: Change of Detection Rate with changes in specificity of match in an immune system simulation
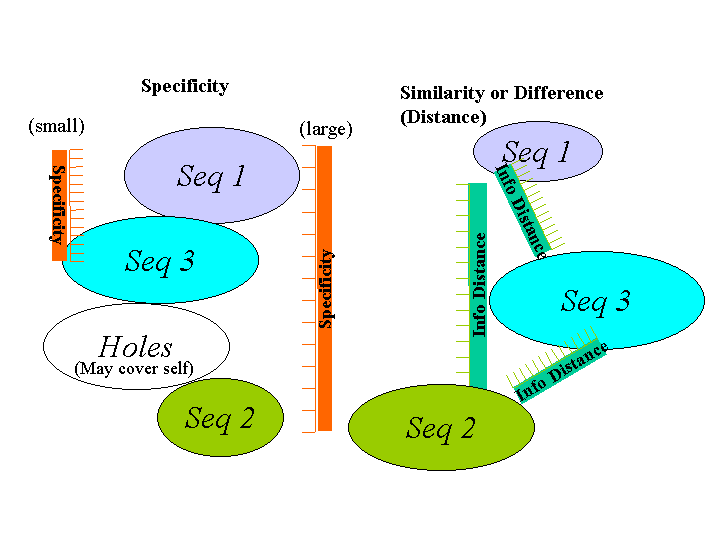
Figure 10: Comparing and Contrasting Specificity and Complexity (Information Distance).
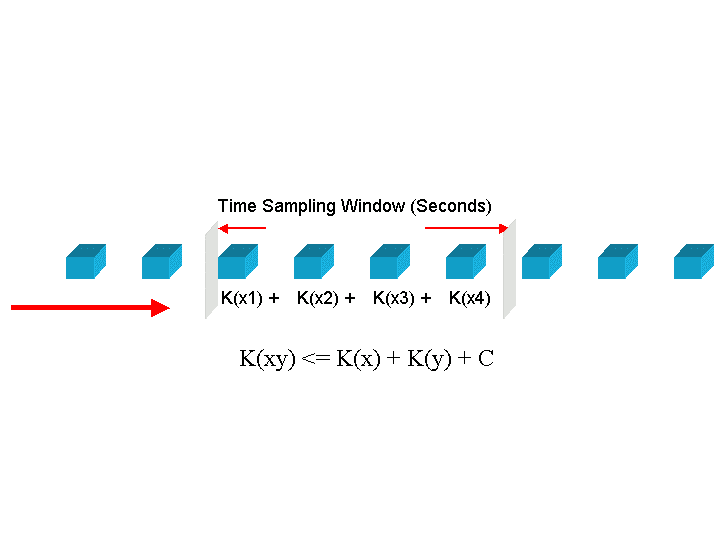
Figure 11: Definition of Differential Complexity.
Table 2: Symbols Used in the Performance Analysis.
| Symbol | Explanation |
|---|---|
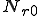 | Number of initial detector strings (before checking for self-match) |
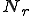 | Number of detector strings after censoring (after removing all self-matches) |
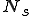 | Number of self-strings |
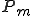 | Probability of match between random strings |
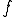 | Probability of a random string not matching any of 'Ns' self strings |
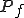 | Probability that 'Nr' detectors fail to detect an intrusion |
 | Length of signature sequence |
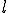 | Length of sequence to be matched (may be longer than signature sequence) |
 | Number of consecutive matches (specificity) |
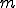 | Number of potential elements at each sequence position; for binary sequences 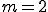 |
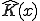 | Kolmogorov Complexity estimate of sequence  |
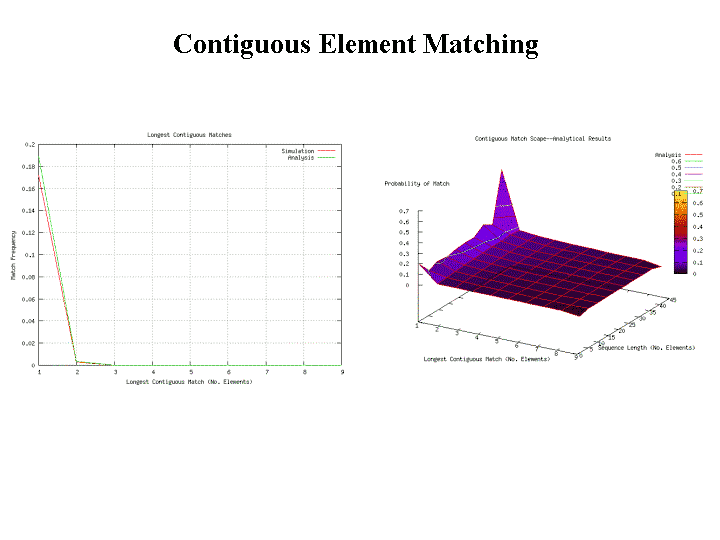
Figure 12: Simulated vs. Analytical Contiguous element matching Match Frequency for 52-Character Strings of Length 10.
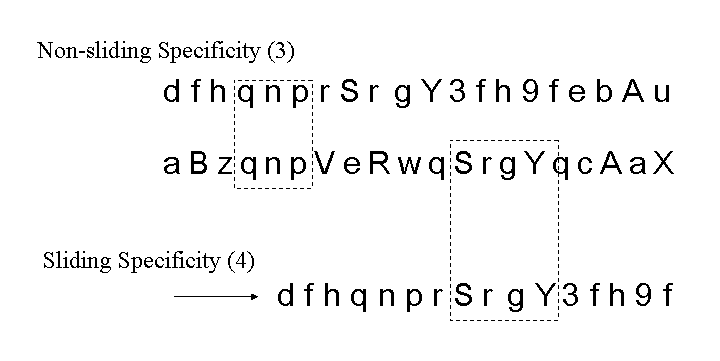
Figure 13: Contiguous Element Matching Returns the Sequence Length of the Longest Number of Contiguous Matches.
![P_m = m^{-r}\left[(1 - r)\frac{(m - 1)}{m} + 1\right]](../img/cache/81722e6455e2742ea7acee8210bb7732.gif)
Equation 3: Probability of Match of r Contiguous Elements in a String.
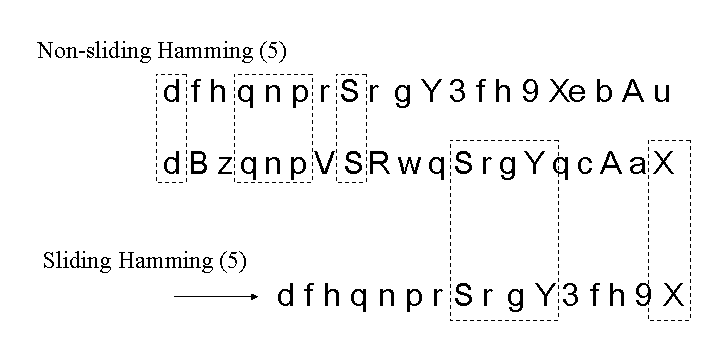
Figure 14: Total Matches Is Maximum Number Of Elements That Match Without The Requirement Of Being Contiguously Located.
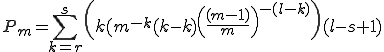
Equation 4: Probability Of Match Of Any Elements In A String Of Sequence Length S And Signature Of Length L.
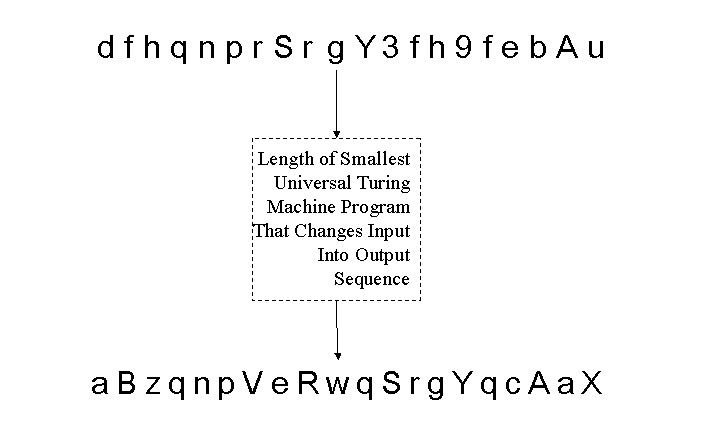
Figure 15: Kolmogorov Complexity is the Length of the Shortest Program that Generates the Signature from the Sequence.
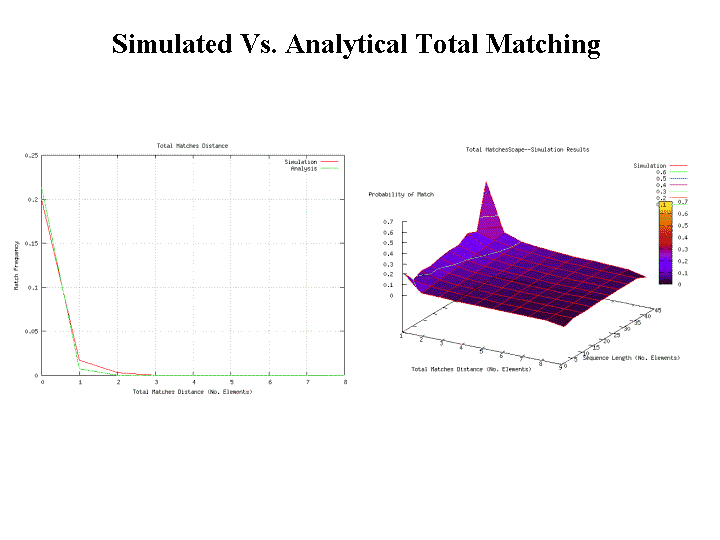
Figure 16: Analytically Derived Probability of Match versus Total Matches and Sequence Length.
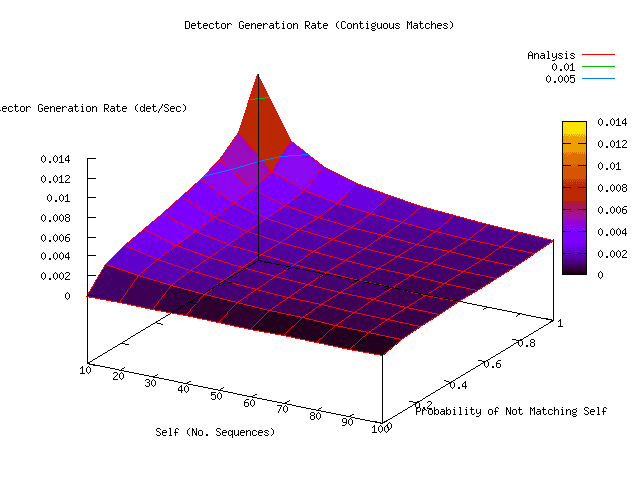
Figure 17: Rate of Detector Generation as a Function of Size of Self and Probability of Matching Self.
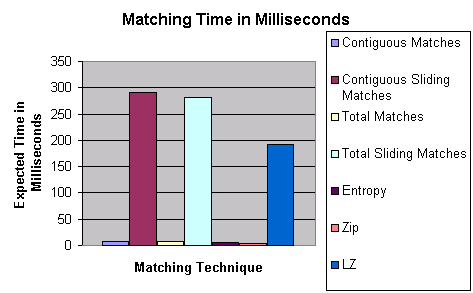
Figure 18: Expected Matching Time for Techniques Discussed in this Paper.
Table 3: Correlation Among Traditional Signature Matching Techniques.
| Contiguous | Sliding Contiguous | Total Matches | Sliding Total Matches | |
|---|---|---|---|---|
| Contiguous | 1.0 | 0.307 | 0.566 | 0.206 |
| Sliding Contiguous | - | 1.0 | 0.179 | 0.521 |
| Total Matches | - | - | 1.0 | 0.335 |
| Sliding Total Matches | - | - | - | 1.0 |
Table 4: Correlation of Complexity Differential Estimates Versus Signature Matching Technique Results.
| Entropy | Zip | LZ | |
|---|---|---|---|
| Contiguous | 0.091 | 0.075 | 0.036 |
| Sliding Contiguous | 0.103 | 0.270 | 0.018 |
| Total Matches | 0.042 | 0.003 | 0.004 |
| Sliding Total Matches | 0.05 | 0.173 | 0.017 |
 Stephen F. Bush (M'03-SM'03) Stephen F. Bush is internationally recognized with over 30 peer-reviewed publications. Dr. Bush has implemented a toolkit capable of injecting predictive models into an active network. The toolkit has been downloaded and used by more than 600 institutions. Stephen continues to explore novel concepts in complexity and algorithmic information theory to refine the toolkit for applications ranging from network management and ad hoc networking to DNA sequence analyses for bioinformatics applications. Dr. Bush has been the Principal Investigator for many DARPA and Lockheed Martin sponsored research projects including: Active Networking (DARPA/ITO), Information Assurance and Survivability Engineering Tools (DARPA/ISO), and Fault Tolerant Networking (DARPA/ATO). Stephen coauthored a book on active network management, titled Active Networks and Active Network Management: A Proactive Management Framework, published by Kluwer Academic Publishers. Before joining GE Global Research, Stephen was a researcher at the Information and Telecommunications Technologies Center (ITTC) at the University of Kansas. He received his B.S. in Electrical and Computer Engineering from Carnegie Mellon University and M.S. in Computer Science from Cleveland State University. He has worked many years for industry in the areas of computer integrated manufacturing and factory automation and control. Steve received the award of Achievement for Professional Initiative and Performance for his work as Technical Project Leader at GE Information Systems
in the areas of network management and control while pursuing his Ph.D. at Case Western Reserve University. Steve completed his Ph.D. research at the University of Kansas where he received a Strobel Scholarship Award. Stephen is currently a Computer Scientist.
Stephen F. Bush (M'03-SM'03) Stephen F. Bush is internationally recognized with over 30 peer-reviewed publications. Dr. Bush has implemented a toolkit capable of injecting predictive models into an active network. The toolkit has been downloaded and used by more than 600 institutions. Stephen continues to explore novel concepts in complexity and algorithmic information theory to refine the toolkit for applications ranging from network management and ad hoc networking to DNA sequence analyses for bioinformatics applications. Dr. Bush has been the Principal Investigator for many DARPA and Lockheed Martin sponsored research projects including: Active Networking (DARPA/ITO), Information Assurance and Survivability Engineering Tools (DARPA/ISO), and Fault Tolerant Networking (DARPA/ATO). Stephen coauthored a book on active network management, titled Active Networks and Active Network Management: A Proactive Management Framework, published by Kluwer Academic Publishers. Before joining GE Global Research, Stephen was a researcher at the Information and Telecommunications Technologies Center (ITTC) at the University of Kansas. He received his B.S. in Electrical and Computer Engineering from Carnegie Mellon University and M.S. in Computer Science from Cleveland State University. He has worked many years for industry in the areas of computer integrated manufacturing and factory automation and control. Steve received the award of Achievement for Professional Initiative and Performance for his work as Technical Project Leader at GE Information Systems
in the areas of network management and control while pursuing his Ph.D. at Case Western Reserve University. Steve completed his Ph.D. research at the University of Kansas where he received a Strobel Scholarship Award. Stephen is currently a Computer Scientist.
 Sanjay Goel is an assistant professor at University at Albany in the School of Business since 2001. Prior to that he has worked for several years in various technical leadership positions at General Electric Corporate Research and Development (now known as GE Global Research Center). Sanjay received his PhD in Mechanical Engineering from Rensselaer Polytechnic Institute in 1998 while working for GE. He also holds a Bachelor of Technology degree in Mechanical Engineering from the Indian Institute of Technology in New Delhi and a Masters degree in the same discipline from Rutgers University. At General Electric he served as a Mechanical Engineer/Software Team Leader, where he developed software for optimization of turbomachinery configuration that was used to design a new class of turbines with improved performance. These products enabled GE to gain a competitive edge in the market. Sanjay also developed a new software tool for automating the design process for steam turbines airfoils that increased designer productivity by five fold. Sanjay has won several management, technical performance and Six Sigma awards, and has authored 19 publications. He is working actively in the area of computer security and peer-to-peer computing. Some of his research projects include: design of computer security systems inspired by biological paradigms of Immunology and Epidemiology, development of self-healing networks based on peer-to-peer architecture, identifying images with hidden data using Information Theory, and use of Machine Learning techniques for reducing complexity of optimization problems. His teaching is focused in the area of Computer Security Networking, Database Design, Enterprise Application Development, and Java Programming. He recently received a $375,000 grant, along with other university faculty, from the Department of Education for development of security curriculum and is currently developing computer security labs in the School of Business. Professor Goel works closely with the New York State Police Computer Crime Unit investigators and the New York Cyber Security and Critical Infrastructure Coordination Agency (CSCIC) in security research and education. He organizes security labs at OSC (Office of the State Comptroller) to provide SUNY students as well as New York State Police Investigators hands-on experience in network hacking and auditing tools.
Sanjay Goel is an assistant professor at University at Albany in the School of Business since 2001. Prior to that he has worked for several years in various technical leadership positions at General Electric Corporate Research and Development (now known as GE Global Research Center). Sanjay received his PhD in Mechanical Engineering from Rensselaer Polytechnic Institute in 1998 while working for GE. He also holds a Bachelor of Technology degree in Mechanical Engineering from the Indian Institute of Technology in New Delhi and a Masters degree in the same discipline from Rutgers University. At General Electric he served as a Mechanical Engineer/Software Team Leader, where he developed software for optimization of turbomachinery configuration that was used to design a new class of turbines with improved performance. These products enabled GE to gain a competitive edge in the market. Sanjay also developed a new software tool for automating the design process for steam turbines airfoils that increased designer productivity by five fold. Sanjay has won several management, technical performance and Six Sigma awards, and has authored 19 publications. He is working actively in the area of computer security and peer-to-peer computing. Some of his research projects include: design of computer security systems inspired by biological paradigms of Immunology and Epidemiology, development of self-healing networks based on peer-to-peer architecture, identifying images with hidden data using Information Theory, and use of Machine Learning techniques for reducing complexity of optimization problems. His teaching is focused in the area of Computer Security Networking, Database Design, Enterprise Application Development, and Java Programming. He recently received a $375,000 grant, along with other university faculty, from the Department of Education for development of security curriculum and is currently developing computer security labs in the School of Business. Professor Goel works closely with the New York State Police Computer Crime Unit investigators and the New York Cyber Security and Critical Infrastructure Coordination Agency (CSCIC) in security research and education. He organizes security labs at OSC (Office of the State Comptroller) to provide SUNY students as well as New York State Police Investigators hands-on experience in network hacking and auditing tools.